Abstract
Eggshell plays an essential role in preventing physical damage and microbial invasions. Therefore, the analysis of genetic regulatory mechanisms of eggshell quality deterioration during aging in laying hens is important for the biosecurity and economic performance of poultry egg production worldwide. This study aimed to compare the differences in the expression profiles of long non-coding RNAs (lncRNAs) and mRNAs between old and young laying hens by the method of high-throughput RNA sequencing to identify candidate genes associated with aging in the uterus of laying hens. Overall, we detected 176 and 383 differentially expressed (DE) lncRNAs and mRNAs, respectively. Moreover, functional annotation analysis based on the Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) databases revealed that DE-lncRNAs and DE-mRNAs were significantly enriched in “phosphate-containing compound metabolic process”, “mitochondrial proton-transporting ATP synthase complex”, “inorganic anion transport”, and other terms related to eggshell calcification and cuticularization. Through integrated analysis, we found that some important genes such as FGF14, COL25A1, GPX8, and GRXCR1 and their corresponding lncRNAs were expressed differentially between two groups, and the results of quantitative real-time polymerase chain reaction (qPCR) among these genes were also in excellent agreement with the sequencing data. In addition, our study found that TCONS_00181492, TCONS_03234147, and TCONS_03123639 in the uterus of laying hens caused deterioration of eggshell quality in the late laying period by up-regulating their corresponding target genes FGF14, COL25A1, and GRXCR1 as well as down-regulating the target gene GPX8 by TCONS_01464392. Our findings will provide a valuable reference for the development of breeding programs aimed at breeding excellent poultry with high eggshell quality or regulating dietary nutrient levels to improve eggshell quality.
1. Introduction
As one of the most affordable sources of available animal protein, eggs are widely favored by consumers around the world, and indeed, eggs dominate commercial markets in many countries [1]. The quality of eggshells, which is both of biological interest and economic importance to the poultry industry, has always been a major concern for the quality and safety of egg products. However, the huge economic loss caused by a deterioration in eggshell quality has been a pressing problem for the egg industry, which has become more acute in the late laying period. The incidence of cracked and broken eggs has been reported to be up to 12–20% in the late laying period, which is one of the key obstacles to extending the laying cycle of laying hens [2]. Most notably, it has been observed that the incidence of damaged and thin-shelled eggs is increased, and the egg production rate is reduced, with the aging of laying hens [3,4]. Therefore, understanding the transcriptomic regulation of eggshell quality with respect to aging is of great economic and biological importance. Further, the deterioration of eggshell quality is directly related to an increased risk of foodborne disease for consumers [5]. Therefore, improving eggshell quality is critically important for the poultry industry and human health.
As is well known, the uterus (shell gland) is composed of glandular and luminal epithelial cells whose secretions promote the biomineralization of the eggshell. The process of eggshell formation is separated into three essential stages: the initiation of crystal growth, linear crystal growth, and the termination of mineralization [6]. An eggshell is a highly ordered structure consisting of a bi-layered membrane (inner and outer), mammillary layer, palisade layer, vertical crystal layer, and cuticle layer. However, eggshell in the late laying period has a lower breaking strength as well as greater variation in structural properties such as thickness, grain morphology, and crystal texture [7]. In addition, the incidence of confluence and early fusion in the mammillary layer decreased with age [8]. Other research has also found that the decreased gland density and a shift in the balance between estrogen receptor α and estrogen receptor β in the shell gland could be critical factors explaining the age-related changes in eggshells [9]. Feng et al. (2020) utilized uterine transcriptome analysis to identify that altered gene expression of matrix proteins in the uterus of laying hens in the late phase of production may trigger age-related impairment of eggshell ultrastructure and its mechanical properties [10]. Therefore, we speculate that deteriorations in eggshell ultrastructure and quality in the late phase of production may be related to age-related abnormalities of gene expression in the uterus. It was supported by the findings that the expression of genes encoding ion transporters and matrix proteins (ATP2A2, SCNN1G, CA2, and ovocalyxin-36) varied with the age of laying hens [11].
Long non-coding RNAs (LncRNAs), with sizes >200 nt, are not translated into proteins [12], are found in both the nucleus and cytoplasm and have received much attention over the past several years. LncRNAs are involved in various aspects of disease and cell and molecular biology, such as cancer, the immune response, neurological and cardiovascular system disorders, cell cycle regulation, cell differentiation, X chromosome inactivation, genomic imprinting, transcriptional control, and epigenetic regulation [13,14,15]. LncRNA, which can regulate target genes in cis and trans, are key regulatory molecules. Cis-acting lncRNAs regulate the expression of target genes that are located at neighboring genomic loci, whereas trans-acting lncRNAs can regulate the expression of transcripts that are located at distal chromosomal loci [16]. The important roles of lncRNAs in the development of different organs and tissue types have been highlighted by many studies. For example, the studies reported here reveal a potential role for the lncRNA MHM and MHM in regulating embryonic growth and gonadal development [17,18]. The loss of lncRNA MHM expression in hens can cause asymmetric development of the ovary, and the loss of lncRNA MHM expression in males may result in decreased expression of the DMRT1 gene in testis [17]. Similarly, the lncRNA alphaGT controls the expression of the α-globin gene from the embryo to the adult and plays a key role in chicken development [19]. However, there is almost no research on the combination of lncRNAs and shell gland development.
To date, however, the potential molecular mechanisms regulating eggshell quality by lncRNAs in the shell gland during aging in laying hens have not been clearly defined. Therefore, in this study, we performed a transcriptomic analysis of the shell gland among old and young laying hens to identify potential mRNAs and lncRNAs that regulate the aging of the shell gland in laying hens. We captured both lncRNAs and mRNAs from fragmented or intact RNA samples to compare whole transcriptomes of old and young chicken shell glands at unprecedented depth. Then, the differentially expressed (DE)-lncRNAs were used in bioinformatics analyses to predict cis- and trans-target genes and to construct lncRNA-mRNA co-expression interaction networks. Next, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analyses were performed to investigate the related roles of differentially expressed genes (DEGs). Our results may help to reveal key mRNAs and lncRNAs involved in the age-related deterioration of eggshell quality and provide potential candidate biomarkers for improving and intervening in eggshell quality in the late laying stage of laying hens.
2. Material and Methods
2.1. Ethics Statement
All the work using animals was approved by the Animal Care and Use Committee of China Agricultural University (No. AW42110202-2-2). All procedures were conducted in accordance with the institutional animal ethics guidelines set by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China.
2.2. Animal and Sample Collection
The eight Hy-Line Brown commercial laying hens used in this study were purchased from Zhuozhou Chicken Farm. These hens were randomly assigned to old (60-week-old, n = 4) and young (31-week-old, n = 4) groups. All birds included in this study were raised on the same diet and managed conditions until slaughtered. Eighteen hours after laying the egg, animals were euthanized with carbon dioxide (approximately 5 min in a small container gassed with carbon dioxide from a compressed gas cylinder). Then, we collected the eggshell glands of each hen from the same pre-determined site and immediately flash frozen in liquid nitrogen and then stored them at −80 °C until RNA extraction.
2.3. Total RNA Extraction
Total RNA was extracted from shell gland tissues using TRIzol reagent according to the manufacturer’s instructions (Invitrogen Life Technologies, Carlsbad, CA, USA). RNA integrity was ascertained by 1.5% agarose gel electrophoresis, and the purity and concentration of the RNA were measured by spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
2.4. cDNA Library Construction and RNA Sequencing
A total of 3 μg RNA per sample was treated with an Epicentre Ribo-zero™ rRNA Removal Kit (Epicentre, Brea, CA, USA) to remove rRNA. The rRNA-free residue was then cleaned up by ethanol precipitation before constructing the RNA-seq libraries. Subsequently, the RNA samples were fragmented and used to synthesize first- and second-strand complementary DNA (cDNA) with random hexamer primers, dNTPs, M-MuLV Reverse Transcriptase (RNaseH-), and DNA Polymerase I. Afterward, the synthetic cDNA fragments were purified using the AMPure XP system (Beckman Coulter, Brea, CA, USA), and the ends were repaired and modified with T4 DNA polymerase and Klenow DNA polymerase to add a single A base and ligate the adapter at the third end of the cDNA fragments. The ligated cDNA products were treated with uracil DNA glycosylase (NEB, Ipswich, MA, USA) to remove the second-strand cDNA. Purified first-strand cDNA was enriched to create the final cDNA library. Lastly, library quality was checked using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). We sequenced the libraries using Illumina HiSeq 2500 Technology (LC Sciences, Houston, TX, USA).
2.5. Sequence Analysis Transcriptome Assembly
Quality control of the RNA-seq reads was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 August 2020)). Clean reads were obtained by removing empty reads, adapter sequences, reads with >10% N sequences, and low-quality reads, in which the number of bases with a quality value Q ≤ 10 was >50%. At the same time, the Q30, GC content, and sequence duplication level of the clean data were calculated. Reads that passed the quality control were then mapped to the Gallus gallus reference genome (Gallus_gallus-5.0). Based on this, the mapped reads of each sample were assembled with StringTie (v1.3.1) using a reference-based approach [20].
2.6. Screening and Prediction of DEGs and DE-lncRNAs
Fragments per kilobase of exon per million fragments mapped (FPKM), means the expected number of fragments per kilobase of transcript sequence per million reads sequenced [21]. It takes into account the effects of sequencing depth and gene length on the fragment count and is currently the most commonly used method for estimating gene expression leve l [22]. In this study, transcript abundance was identified by FPKM using Cuffdiff (http://cufflinks.cbcb.umd.edu/manual.html#cuffdiff (accessed on 28 August 2020)) [23]. Here, FPKM was used to calculate the fold change of DEGs between the two groups, and the FPKM of the protein-coding genes in each sample was computed by summing the FPKMs of the transcripts in each gene group. Moreover, we analyzed DEGs by using the edgeR package to calculate the p-value that was obtained by multiple hypothesis testing calibrations [24,25]. LncRNAs or protein-coding genes with p < 0.05 and log2 (fold change) > 1 were assigned as DEGs.
2.7. Construction of the LncRNA-Gene Interaction Network
Previous studies confirm that lncRNAs can regulate gene expression through cis-acting and trans-acting mechanisms [26]. For each lncRNA locus, the 10 k/100 k upstream and downstream protein-coding genes (without overlap) were first identified as cis-target genes. However, the genes that overlapped with the lncRNAs predicted by Lnctar (http://www.cuilab.cn/lnctar (accessed on 12 October 2020)) were selected as trans-target genes. To further investigate the interactions between the DE-lncRNAs and their corresponding differentially expressed cis- or trans-target genes, we constructed an interactive lncRNA-gene network based on their FPKM using Cytoscape 3.8.2 software (http://www.cytoscape.org (accessed on 22 October 2020)). Moreover, we calculated the Pearson correlation coefficient (COR) of each lncRNA and DEG expression value.
2.8. GO and Pathway Analysis
GO enrichment analysis of DEGs or lncRNA target genes was implemented using the Molecule Annotation System (MAS) 3.0 (http://bioinfo.capitalbio.com/mas3 (accessed on 2 November 2020)), which is based on the KEGG database (Capital Bio, Beijing). GO terms with p < 0.05 were considered significantly enriched by DEGs.
KEGG is a database resource for understanding high-level functions and utilities of a biological system [27], such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/ (accessed on 2 November 2020)). We used KOBAS 3.0 software to test the statistical enrichment of DEGs or lncRNA target genes in KEGG pathways [28].
2.9. Analysis of the Expression Levels and Validation by qPCR
For validation via the quantitative real-time polymerase chain reaction (qPCR), single-stranded cDNA was synthesized from 1 µg of total RNA in a final volume of 20 μL according to the manufacturer’s protocol (PrimeScriptTM RT reagent Kit with gDNA Eraser, TaKaRa, Dalian, China). The qPCR reactions were performed on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) in a 20 μL volume using Fast Start Universal SYBR Green Master (ROX) (TaKaRa, Dalian, China), and each sample was analyzed in triplicate. The cycling conditions were 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 34 s. A melting curve was obtained at 60–95 °C for each sample amplified. In this study, qPCR primers were designed using the Premier Primer 5.0 software (Premier Biosoft International, San Francisco, CA, USA) and the sequences in GenBank (https://www.ncbi.nlm.nih.gov/ (accessed on 12 February 2021)) and from RNA-seq. The chicken β-actin gene was used as an internal control. The qPCR primer sequences are presented in Table 1.

Table 1.
Primer sequences of qPCR.
2.10. Statistical Analysis
The results of quantitative expression are presented as the mean ± standard error (SEM), and the significance of the data was tested by a two-tailed paired Student’s t-test using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA). The 2−ΔΔCt method was used to analyze the results of qPCR as described [29], and β-actin was used as an internal control to normalize all of the threshold cycle (Ct) values.
3. Results
3.1. Reads Mapping
In total, we obtained 82,871,160–86,696,604 and 86,622,130–86,688,990 raw reads from the libraries of shell gland tissues of old chickens (n = 4) and young chickens (n = 4), respectively. Correspondingly, we ultimately obtained 80,510,552–138,847,948 and 84,918,798–85,469,778 clean reads by filtering and removing sequence reads with adapters and low quality, respectively. In addition, the Q30 of each sample was not <90.85%. The quality value of Q30 indicated a 0.1% probability of error base calling during sequencing. Moreover, it was generally accepted that the number of bases with a base quality above Q30 was more than 85%, indicating that the sequencing quality of each sample was high and met the requirements for library construction. Subsequently, we found that >78.77% of the clean reads were completely mapped to the chicken reference genome. The unique mapped reads ranged from 66.24–80.46% of the total mapped reads (Table 2).

Table 2.
Summary of clean reads mapping to the chicken reference genome.
3.2. Identification and Characterization of LncRNAs
We performed a comparative analysis of the structure of lncRNAs and mRNAs to study the basic features of lncRNAs in the chicken shell gland. This was not just to determine the difference between lncRNAs and mRNAs but also to verify if the predicted lncRNAs were consistent with general characteristics. In this study, the intersection of the Coding Potential Calculator (CPC), Coding-Non-Coding Index (CNCI), and Protein Families Database (PFAM) results yielded 5334 lncRNA transcripts, including the identified conservative lncRNAs (Figure 1A). Interestingly, previous reports indicate that protein-coding transcripts are longer and more conserved than lncRNAs [23]. In agreement with this, we found that the predicated lncRNAs are shorter in length than protein-coding transcripts (Figure 1B) and tend to contain fewer exons (Figure 1C). We also found that the average open reading frame (ORF) length of the predicted lncRNAs was 126 amino acids (aa), which was less than mRNA (687 aa, Figure 1D).
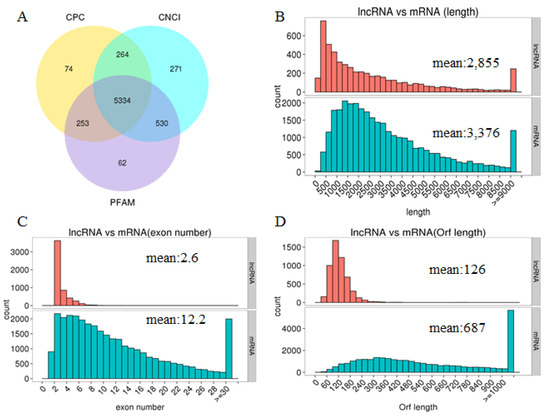
Figure 1.
The features of predicted lncRNAs and mRNAs. (A) Venn diagram of lncRNAs from the Coding Potential Calculator (CPC), the Coding-Non-Coding Index (CNCI), and Protein Families Database (PFAM). (B) Length distribution of lncRNAs and coding transcripts. (C) Exon number distribution of lncRNAs and coding transcripts. (D) Orf length distribution of lncRNAs and coding transcripts.
3.3. Differential Expression of Predicted LncRNAs and mRNAs in the Eggshell Gland
The expression level of each lncRNA and mRNA was estimated by FPKM using Cuffdiff. To explore similarities and to compare the relationships between the different libraries, we measured the expression patterns of DE-lncRNAs and protein-coding genes by systematic cluster analysis (Figure 2). As a result, we identified 176 lncRNA transcripts that were expressed differentially in the eggshell glands between the old group and young group (Supplementary Table S1), and the sequences could be found in the Supplementary File S1. Compared to the young group, 91 lncRNAs were up-regulated, and 85 lncRNAs were down-regulated, in the old group. Among these, the 20 most significantly up-regulated or down-regulated lncRNAs are presented in Table 3 (Figure 2A,B and Table 3).
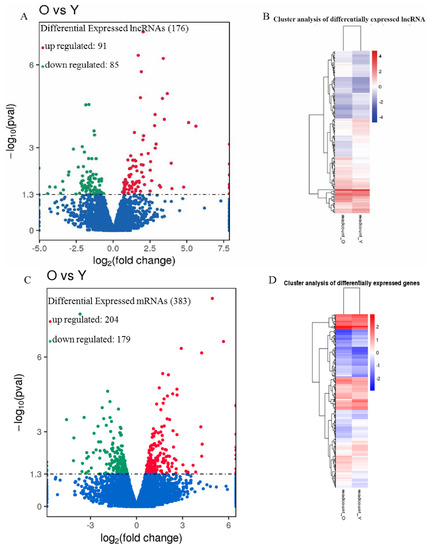
Figure 2.
Analyses of DE-lncRNAs and mRNAs in the eggshell gland. (A) The volcano plot can intuitively see the overall distribution of the differential transcripts, and the threshold value was set to p < 0.05. Blue dots represent that lncRNAs are not significantly differential expressions; Red dots represent relatively high expressions; Green dots represent relatively low expressions. (B) A Heatmap of 176 lncRNA expression profiles showed significant expression differences (91 up-regulated and 85 down-regulated). Data were expressed as FPKM, and the red-to-green color gradient indicates from high expression to low expression. (C) The volcano plot can intuitively see the overall distribution of the differential genes, and the threshold value was set to p < 0.05. Blue dots represent that lncRNAs are not significantly differential expressions; Red dots represent relatively high expressions; Green dots represent relatively low expressions. (D) The Heatmap of 383 mRNAs expression profiles showed significant expression differences (204 up-regulated and 179 down-regulated). Data were expressed as FPKM, and the red-to-green color gradient indicates from high expression to low expression.

Table 3.
The top 20 up-regulated or down-regulated lncRNAs.
Differential expression of mRNAs in shell gland tissues of the old group was also compared to that in the young group. A total of 383 mRNAs were found to be expressed differentially, with 204 up-regulated and 179 down-regulated (Figure 2C,D and Supplementary Table S2).
3.4. Construction of the LncRNA-mRNA Co-Expression Network
To investigate the questions of whether the functions of DE-lncRNAs are in agreement with their target genes in regulating the chicken eggshell gland, and how do lncRNAs and their target genes interact (cis or trans), we constructed a co-expression network between DE-lncRNAs and their significantly correlated DE cis- and trans-target genes using Cytoscape (Figure 3). For the old chicken vs. young chicken comparison, the lncRNA-mRNA co-expression interaction network comprised 37 network nodes and 48 lncRNA-gene connections among 13 DE-lncRNAs and 24 DE-mRNAs. Further analysis revealed that lncRNA upregulated in the shell gland of old chickens increased the expression of cis- and trans-target genes, except for upregulated TCONS_03323652 which decreased the cis-target gene LOC422895. In particular, TCONS_00181492 and TCONS_03123639 showed a significant positive correlation with the cis-target genes FGF14 and GRXCR1, which are closely associated with calcium and sodium plasma transport [30,31]. In addition, calcium ion transport has a vital role in the eggshell formation, suggesting that TCONS_00181492 and TCONS_03123639 play an essential role in regulating eggshell formation.
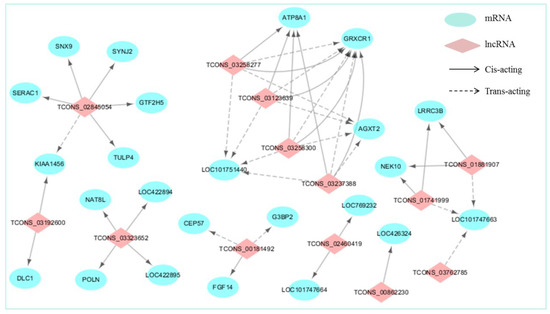
Figure 3.
LncRNAs-mRNAs co-expression interaction network. DE-lncRNAs (p-adjust < 0.05) and their corresponding differentially expressed cis- and trans-target genes (p-adjust < 0.05) were selected and used to construct a lncRNAs-mRNAs co-expression network. In this network, protein-coding genes are displayed as blue circles, and lncRNA are displayed as pink diamonds. Solid lines mean the interactions between DE-lncRNAs and their corresponding cis-target genes, whereas the dashed lines mean interactions between DE-lncRNAs and their corresponding trans-target genes.
3.5. Enrichment Analysis of the Nearest Neighbor Genes of the lncRNAs
To investigate the functions of the lncRNAs, we predicted their potential cis targets. We searched for protein-coding genes 10 kb and 100 kb upstream and downstream of all of the identified lncRNAs. We found 176 lncRNAs that were transcribed close to (<10 kb) 206 neighboring protein-coding genes, and 176 lncRNAs that were transcribed close to (<100 kb) 154 neighboring protein-coding genes (Supplementary Tables S3 and S4). To explore the functions between lncRNAs and their cis-regulated target genes, we performed GO analysis. We found 90 GO terms (<10 kb) that were significantly enriched (p < 0.05) (Supplementary Table S5), and most of these terms were associated with biological processes and molecular functions (Supplementary Figure S1). In addition, we found 140 GO terms (<100 kb) that were significantly enriched (p < 0.05) (Supplementary Table S6), and most of these terms were associated with biological processes, molecular functions, and cellular components (Supplementary Figure S2). For example, the main enriched terms included “protein phosphorylation (GO:0006468)”, “phosphate-containing compound metabolic process (GO:0006796)”, “phosphorus metabolic process (GO:0006793)”, “protein modification process (GO:0006464)”, “ATP binding (GO:0005524)”, “ATP-dependent helicase activity (GO:0008026)”, and “mitochondrial proton-transporting ATP synthase complex (GO:0005753)” (Table 4 and Table 5). Most of them were closely related to the formation of eggshells, which suggests that one of the principal roles of lncRNAs may be to regulate the synthesis and metabolism of organics and minerals. Pathway analysis indicated that cis-target genes were significantly enriched in four (<10 kb) and six (<100 kb) KEGG pathways (p < 0.05), respectively (Table 6 and Table 7). These data suggest that the function of the shell gland may be regulated by the action of lncRNAs on these neighboring protein-coding genes.

Table 4.
Gene Ontology (GO) enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<10 kb) lncRNAs (GO level > 3).

Table 5.
Gene Ontology (GO) enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<100 kb) lncRNAs (GO level > 3).

Table 6.
KEGG pathway enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<10 kb) lncRNAs (p < 0.05).

Table 7.
KEGG pathway enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<100 kb) lncRNAs (p < 0.05).
3.6. Enrichment Analysis of Co-Expressed Genes of lncRNAs
We also predicted the potential targets of lncRNAs in trans-regulatory relationships using co-expression analysis. The COR method was used to analyze the correlation between the lncRNAs and mRNAs in samples, and the main functions of the lncRNAs were predicted using mRNA, with a correlation absolute value >0.95. We found 176 lncRNAs that were transcribed close to 791 protein-coding genes (Supplementary Table S7). Functional analysis indicated that the co-expressed genes were significantly enriched in 174 GO terms (95 under biological process, 43 under cellular component, and 36 under molecular function) that encompassed a variety of biological processes (p < 0.05) (Supplementary Table S8 and Figure S3). Importantly, some of the terms were related to organic metabolism and genetic development, including “cellular protein metabolic process (GO:0044267)”, “macromolecule biosynthetic process (GO:0009059)”, “protein metabolic process (GO:0019538)”, “Ras GTPase binding (GO:0017016)”, and “GTPase binding (GO:0051020)” (Table 8). Most of them were associated with organic synthesis and metabolism. The co-expressed genes were significantly enriched in nine KEGG pathways (p < 0.05) (Table 9), where the pathways “Salmonella infection” and “AGE-RAGE signaling pathway in diabetic complications” affected the function of the shell gland of aging laying hens. As the disease resistance of aging hens is weakened, the metabolism and synthesis ability of the body is reduced, which leads to a decline in eggshell quality.

Table 8.
Gene Ontology (GO) enrichment analysis of differentially expressed protein-coding genes targeted by trans-acting lncRNAs (GO level > 3).

Table 9.
KEGG pathway enrichment analysis of differentially expressed protein-coding genes targeted by trans-acting lncRNAs (p < 0.05).
3.7. Enrichment Analysis of DE-mRNAs
To further understand the biological processes regulated during eggshell formation and to determine which processes are encoded by DEGs, we performed GO and KEGG enrichment analyses on 383 mRNAs. We found 124 GO terms that were significantly enriched (p < 0.05) (Supplementary Table S9), and most of these terms were associated with biological processes, molecular function, and cellular components (Table 10 and Supplementary Figure S4). The majority of DEGs were categorized as ion transport in the eggshell gland during the formation of the eggshell. The GO terms included “inorganic anion transport (GO:0015698)”, “inorganic anion transmembrane transporter activity (GO:0015103)”, “anion transmembrane transporter activity (GO:0008509)”, “electron carrier activity (GO:0009055)”, and “calcium ion binding (GO:0005509)”. Thirty-one genes were categorized under these terms, including Glutaredoxin cysteine-rich 1 (GRXCR1), and the members (SLC1A3, SLC6A4, SLC20A1, SLC22A13, SLC26A3, SLC30A8, SLC39A2, SLC43A3, and SLC45A2) of the sodium-dependent phosphate transporter family. GO term analysis also revealed some DEGs with possible roles in protein translation and binding. The terms included “protein polymerization (GO:0051258)”, “regulation of G-protein coupled receptor protein signaling pathway (GO:0008277)”, “transcription factor complex (GO:0005667)”, “DNA-directed RNA polymerase II, holoenzyme (GO:0016591)”, and “protein binding, bridging (GO:0030674)”. Several genes were enriched in these terms, most notably FGF14 and COL5A2. Another important group of DEGs were involved in membrane fiber formation and/or encoded extracellular matrix proteins; “extracellular space (GO:0005615)”, “membrane (GO:0016020)”, and “fibrinogen complex (GO:0005577)” were implicated in this.

Table 10.
Gene Ontology (GO) enrichment analysis of DE-mRNAs (GO level > 3).
In addition, we also found 10 KEGG pathways that were significantly enriched (p < 0.05) (Table 11), several of which were related to the function of the shell gland, including “Glycine, serine, and threonine metabolism”, “ABC transporters”, and “Toll-like receptor signaling pathway”. The D-3-phosphoglycerate dehydrogenase (PHGDH) gene was significantly enriched in the serine metabolism pathway, and the osteopontin (SPP1) gene is a matrix protein that was significantly enriched in the “Toll-like receptor signaling pathway”.

Table 11.
KEGG pathway enrichment analysis of DE-mRNAs (p < 0.05).
3.8. Validation of DE-lncRNAs and -mRNAs
To further validate the reliability and reproducibility of our RNA-seq data, four DE-lncRNAs (TCONS_00181492, TCONS_03234147, TCONS_03123639, and TCONS_01464392) and their corresponding target genes (FGF14, COL25A1, GRXCR1, and GPX8) related to eggshell quality were randomly selected for qPCR validation. The analysis showed that the expression tendencies of all four lncRNAs and their target genes were extremely concordant with the RNA-seq data, though the absolute fold changes differed between qPCR and RNA-seq (Figure 4 and Supplementary Table S10). Appreciably, TCONS_00181492, TCONS_03234147, and TCONS_03123639 up-regulated their corresponding target genes, but TCONS_01464392 down-regulated GPX8. These results are consistent with that of the co-expression interaction network, especially for TCONS_00181492 and TCONS_03123639 regulating FGF14 and GRXCR1, respectively.
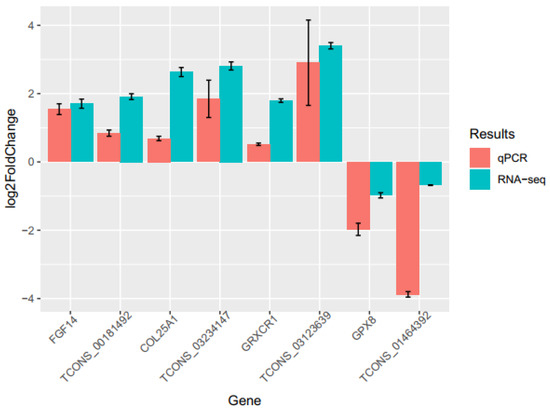
Figure 4.
Validation of 4 DE-lncRNAs and their target genes by qPCR. The qPCR results of the DE-lncRNAs and DEGs were compared with their RNA-Seq, respectively. Red: represents qPCR, and Blue: represents RNA-seq.
4. Discussion
Comparative transcriptome analyses of organs or tissues at different developmental stages can provide valuable insights into the question of how regulatory gene interaction networks control specific biological processes and how diseases can arise [32]. Recently, increasing evidence has confirmed that lncRNAs are important regulatory factors of gene expression, regulating target genes by cis-acting (neighboring genes) or trans-acting (distant genes) mechanisms [33]. Furthermore, RNA-seq has been performed to provide an extensive lncRNA and gene expression profile in different tissues of livestock and poultry (e.g., cell differentiation and development [34], cancer [35], and skeletal muscle development [36]. Previous studies of the hen uterus transcriptome and gene expression profiling during the formation of the eggshell demonstrate a large number of DEGs that participate in ion transport for eggshell mineralization and the secretion of matrix proteins [37,38,39,40,41]. Most of the previous studies report the roles of mRNAs in the avian eggshell gland, but systematic identification of the functions of lncRNAs remained unclear in the development of the chicken shell gland. Therefore, in this study, we performed transcriptome sequencing of the shell gland of laying hens in the peak and late laying periods and analyzed the DE-lncRNAs and DEGs to reveal their roles in eggshell quality. To the best of our knowledge, this study represents the first systematic genome-wide analysis of lncRNAs and mRNAs in the chicken shell gland, providing a valuable catalog of functional lncRNAs and mRNAs associated with eggshell quality.
In the present study, we developed a highly stringent filtering pipeline to minimize the selection of false positive lncRNAs, which aimed to remove transcripts with evidence of protein-coding potential, and performed co-location mRNA prediction and co-expression mRNA prediction for the lncRNAs obtained from the chicken eggshell gland. Ultimately, we identified 176 DE-lncRNAs and 383 DE-mRNAs. To gain insight into how interactions between DE-lncRNAs and their corresponding target genes regulate shell gland development, we constructed co-expression interaction networks between DE-lncRNAs and their predicted cis- and trans-target genes. Then, four DE-lncRNAs and their target genes related to eggshell quality were selected for qPCR validation, and the results were consistent with the RNA-seq data, which demonstrated that lncRNA TCONS_01464392 can target the GPX8 gene, and they are all down-regulated. LncRNAs TCONS_00181492, TCONS_03234147, and TCONS_03123639 target FGF14, COL25A1, and GRXCR1, respectively, and these six genes are up-regulated.
The oviduct of hens is composed of the infundibulum, magnum, isthmus, shell gland, and vagina. Especially, the shell gland is the place where the eggshell is deposited [42]. The formation of the eggshell is a complex process involving the precipitation of calcium carbonate [43]. Mature follicles reach the shell gland and calcify layer by layer. After the mature follicles reach the shell gland, they need to go through the calcification process, and eventually form the eggshell, and the whole process takes about 15–16 h. Approximately 94% of minerals in the eggshell are calcium carbonate, with other inorganic minerals being calcium phosphate, magnesium phosphate, and magnesium carbonate [43]. Previous studies suggest that eggshell calcification requires the interaction of numerous processes, including transcellular and paracellular transport of minerals and the secretion of different matrix proteins [44,45,46]. Particularly, ion transportation plays a crucial role in the process of eggshell formation. The ion channels contribute to the transportation of Ca2+ from the plasma to the uterine lumen, which includes Na+, Ca2+, and K+ channels [47]. Moreover, the characteristics of eggshell calcification in poultry are that the body rapidly and massively transports Ca2+ from blood to the lumen of the eggshell gland, and a calcium ATPase (calcium pump) is a key enzyme involved in Ca2+ transport in the uterus during eggshell formation [48]. Apart from Ca2+, inorganic phosphate (Pi) is also essential in the formation of eggshells. Pi is involved in many biological processes, including nucleic acid synthesis, skeletal development, signaling cascades, and tooth mineralization [49,50,51]. More meaningfully, phosphorus participates in the transport mechanism of the calcium pump (calcium ATPase).
In the present study, we conducted GO and KEGG pathway enrichment analyses on DE-mRNAs and DE-lncRNAs and found that the most of identified DEGs were involved in eggshell calcification and cuticularization pathways, such as “inorganic anion transport”, “inorganic anion transmembrane transporter activity”, “phosphate-containing compound metabolic process”, “phosphorus metabolic process”, “protein metabolic process”, “mitochondrial proton-transporting ATP synthase complex”, “proton-transporting ATP synthase complex”, and “calcium ion binding”. Notably, SPP1 was significantly enriched in the “Toll-like receptor signaling pathway”, and the authors of a previous study suggest that SPP1 is differentially expressed in the uterus between a low eggshell strength group and a normal eggshell strength group during eggshell formation [41]. In addition, another study indicates that the PHGDH gene is highly over-expressed in the isthmus during the deposition of the eggshell membranes [40]. The PHGDH gene was also differentially expressed between two groups and enriched in the “Glycine, serine, and threonine metabolism pathway” in this study. Hence, we speculate that the deterioration of eggshell quality during aging in laying hens may be due to disruption of inorganic ion and amino acid transport in the shell gland.
Based on the lncRNA-mRNA co-expression interaction networks, the predicted target gene of lncRNA TCONS_00181492 is FGF14. Prior to this analysis, little was known concerning the association between FGF14 and lncRNA. FGF14 is a well-known growth factor belonging to the FGF family. FGF family members possess broad mitogenic and cell survival activities and are involved in a variety of biological processes, including cell growth, embryonic development, tissue repair, morphogenesis, tumor growth, and invasion [52]. Previous work demonstrates that FGF14 is a functionally relevant component of the neuronal voltage-gated Na+ (Nav) channel complex [53], and FGF14 can also regulate members of the presynaptic Cav2 Ca2+ channel family [54]. Simultaneously, there is evidence that the transfer and concentration of Na+ can directly affect the transportation of Ca2+ and HCO3− in the chicken uterus [30]. In the present study, we found that the expression of FGF14 is up-regulated in the shell gland of chickens in the old group as compared to the young group. The aforementioned studies indicate that the FGF14 gene plays an important role in chicken growth [31]. The predicted regulatory lncRNA, TCONS_00181492, was significantly more highly expressed in the shell gland in the old group than in the young group and controlled the expression of FGF14 via cis-acting mechanisms. Furthermore, TCONS_00181492 and FGF14 were positively correlated. Therefore, we had reason to speculate that TCONS_00181492 may regulate shell gland development in the chicken via the cis-acting target gene FGF14. Thus, we conjecture that the expression of the target gene FGF14 may be regulated by TCONS_00181492 through cis-acting in the shell gland during the aging of laying hens, thereby affecting the ion transport in the shell gland and ultimately leading to the deterioration of eggshell quality.
COL25A1 was a predicted cis-target of TCONS_03234147 that is related to the focal adhesion pathway. Collagen XXV α 1 (COL25A1), the extracellular matrix gene, is a collagenous type II transmembrane protein, which was first purified from senile plaques of Alzheimer’s disease (AD) brains [55]. In recent years, work on collagen genes has attracted the attention of many researchers. Previous studies of the hen oviduct transcriptome during eggshell membrane formation identify a large number of differentially expressed collagen genes, such as collagen X (COL10A1), collagen I (COL1A1), collagen II (COL2A1), and collagen III (COL3A1) [40]. Moreover, COL11A1 was also differentially expressed between the normal eggshell strength group and low eggshell strength group in the study integrating transcriptome and genome re-sequencing in the chicken uterus [41]. TCONS_03234147 and its target gene COL25A1 were differentially expressed between the two groups in the present study, and their expression was higher in aging hens compared to young hens.
The GRXCR1 gene is the putative cis-target of TCONS_03123639 in the lncRNAs-genes network. The GRXCR1 gene encodes an evolutionarily conserved cysteine-rich protein with sequence similarity to the glutaredoxin family of proteins [56]. Recently, research on the function of the GRXCR1 gene has mostly been focused on diseases [57]. However, the biological function of the GRXCR1 gene is still rarely reported in livestock and poultry research. Herein, we found that GRXCR1 was enriched in the ion transport pathway, implying that GRXCR1 may play an important role in the formation of eggshells. Remarkably, the members (SLC1A3, SLC6A4, SLC20A1, SLC22A13, SLC26A3, SLC30A8, SLC39A2, SLC43A3, and SLC45A2) of the sodium-dependent phosphate transporter (SLC) family were also enriched in ion transport pathways (Table 10). Previous studies show that zinc ion transporters include two major families, SLC30 (Solute-Linked Carrier30, also named ZnT) and SLC39 (Solute-Linked Carrier 39, also named ZIP). ZnT contains 10 transporters of SLC30A1-SLC30A10, and ZIP contains 14 transporters of SLC39A1-SLC39A14. In our study, the differentially expressed SLC30A8 and SLC39A2 genes belong to the ZnT family and ZIP family, respectively. Carbonic anhydrase located in the eggshell gland epithelial cells is an important enzyme in the process of eggshell formation, which can reversibly catalyze the hydrolysis of H2CO3, regulate the concentration of HCO− in the eggshell gland, and then affect the Ca2+ transport process and the calcium deposition in the eggshell, changing the quality of the eggshell. Zinc ions are necessary for the activity center of carbonic anhydrase, so zinc can affect the activity of carbonic anhydrase [58]. Moreover, zinc is also a component of alkaline phosphatase, which may regulate some phosphorylated proteins related to the mechanism of eggshell formation and affect the synthesis of calcium carbonate crystals [59]. This provides us a vision for adding appropriate zinc to the diet of aging laying hens, which may reduce the deterioration of eggshell quality.
Through integration analysis of bioinformatics, we found that the differentially expressed TCONS_01464392 could target the GPX8 gene, whose expression was extremely significant, and their expression levels were negatively correlated. Glutathione peroxidases (GPXs) are enzymes that are present in almost all organisms, with the primary function of limiting peroxide accumulation. In mammals, GPXs consist of eight isoforms, but only two members (GPX7 and GPX8) reside in the endoplasmic reticulum [60,61]. A previous study demonstrates that GPX8 is enriched in mitochondria-associated membranes and can regulate Ca2+ storage and fluxes [61]. This indicates that the decline in eggshell quality of aging laying hens may be closely related to down-regulated GPX8 expression levels.
In conclusion, this research presents the first analysis of lncRNA and mRNA expression in the uterus during the aging of laying hens. A total of 176 DE-lncRNAs and 383 DEGs were identified by comparing the uterus of old and young laying hens. Several novel lncRNAs were revealed. Moreover, functional annotation analysis based on the Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) databases revealed that DE-lncRNAs and DE-mRNAs were significantly enriched in “phosphate-containing compound metabolic process”, “mitochondrial proton-transporting ATP synthase complex”, “inorganic anion transport”, and other terms related to eggshell calcification and cuticularization. These results suggested that lncRNAs in the uterus regulated eggshell quality during aging in laying hens by targeting key genes that modulate ion transport and eggshell calcification. Eight highly associated genes were identified and validated by RT-qPCR, and the results were consistent with the RNA-seq results. These findings provide a solid foundation for future studies on the molecular mechanisms of oviductal senescence in laying hens. The findings laid a solid foundation for future studies on the molecular mechanisms of oviduct aging of laying hens. These findings provide a solid foundation for future studies on the molecular mechanisms of oviduct aging and improve eggshell quality in late-laying hens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14030639/s1, Figure S1: GO enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<10 kb) lncRNAs. Figure S2: Gene Ontology (GO) enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<100 kb) lncRNAs. Figure S3: GO enrichment analysis of differentially expressed protein-coding genes targeted by trans-acting lncRNAs. Figure S4: GO enrichment analysis of differentially expressed protein-coding genes. Table S1: Differential expression of lncRNAs in eggshell gland tissues. Table S2: Differential expression of mRNAs in eggshell gland tissues. Table S3: LncRNAs and their cis-regulated (<10 kb) target genes. Table S4: LncRNAs and their cis-regulated (<100 kb) target genes. Table S5: GO enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<10 kb) lncRNAs. Table S6: GO enrichment analysis of differentially expressed protein-coding genes targeted by cis-acting (<100 kb) lncRNAs. Table S7: LncRNAs and their trans-regulated target genes. Table S8: GO enrichment analysis of differentially expressed protein-coding genes targeted by trans-acting lncRNAs. Table S9: Gene Ontology (GO) enrichment analysis of differentially expressed protein-coding genes. Table S10: Validation of RNA-seq results by using qPCR. File S1: >TCONS_04570322.
Author Contributions
Conceptualization, G.L and J.L.; writing—original draft preparation, G.L.; writing—review and editing, B.Z. and J.L.; data curation, X.Y. and G.L., visualization, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2022YFE0111100) and by the 2115 Talent Development Program of China Agricultural University.
Institutional Review Board Statement
The study was approved by the Ethics Committees of Laboratory Animal Center of China Agricultural University. All experimental procedures were performed according to protocols approved by the Institutional Studies Animal Care and Use Committee of China Agricultural University (Beijing, China).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare that no conflict of interest exists.
Abbreviations
| LncRNAs | long non-coding RNAs |
| RNA-seq | RNA sequencing |
| DE | differentially expressed |
| qPCR | the quantitative real-time polymerase chain reaction |
| DEGs | differentially expressed genes |
| FPKM | fragments per kilobase of exon per million fragments mapped |
| COR | Pearson correlation coefficient |
| Orf | open reading frame |
| O | old group |
| Y | young group |
References
- Mottet, A.; Tempio, G. Global poultry production: Current state and future outlook and challenges. World’s Poult. Sci. J. 2017, 73, 1–12. [Google Scholar] [CrossRef]
- Travel, A.; Nys, Y.; Bain, M.M. Effect of hen age, moult, laying environment and egg storage on egg quality. In Improving the Safety and Quality of Eggs and Egg Products; Nys, Y., Bain, M., van Immerseel, F., Eds.; Woodhead Publishing Limited: Cambridge, UK, 2011; pp. 300–329. [Google Scholar]
- Joyner, C.J.; Peddie, M.J.; Taylor, T.G. The effect of age on egg production in the domestic hen. Gen. Comp. Endocrinol. 1987, 65, 331–336. [Google Scholar] [CrossRef] [PubMed]
- AlBatshan, H.A.; Scheideler, S.E.; Black, B.L.; Garlich, J.D.; Anderson, K.E. Duodenal Calcium Uptake, Femur Ash, and Eggshell Quality Decline with Age and Increase Following Molt. Poult. Sci. 1994, 73, 1590–1596. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Guo, Y.; Li, W.; Song, J.; Xu, G.; Yang, N.; Zheng, J. Impact of cuticle quality and eggshell thickness on egg antibacterial efficiency. Poult. Sci. 2019, 98, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Marie, P.; Labas, V.; Brionne, A.; Harichaux, G.; Hennequet-Antier, C.; Nys, Y.; Gautron, J. Quantitative proteomics and bioinformatic analysis provide new insight into protein function during avian eggshell biomineralization. J. Proteom. 2015, 126, 140–154. [Google Scholar] [CrossRef]
- Athanasiadou, D.; Jiang, W.; Goldbaum, D.; Saleem, A.; Basu, K.; Pacella, M.S.; Böhm, C.F.; Chromik, R.R.; Hincke, M.T.; Rodríguez-Navarro, A.B.; et al. Nanostructure, osteopontin, and mechanical properties of calcitic avian eggshell. Sci. Adv. 2018, 4, eaar3219. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.R.; Chousalkar, K.K. Effect of production system and flock age on egg quality and total bacterial load in commercial laying hens. J. Appl. Poult. Res. 2014, 23, 59–70. [Google Scholar]
- Wistedt, A.; Ridderstrale, Y.; Wall, H.; Holm, L. Age-related changes in the shell gland and duodenum in relation to shell quality and bone strength in commercial laying hen hybrids. Acta. Vet. Scand. 2019, 61, 14. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, H.J.; Wu, S.G.; Qi, G.H.; Wang, J. Uterine transcriptome analysis reveals mRNA expression changes associated with the ultrastructure differences of eggshell in young and aged laying hens. BMC Genom. 2020, 21, 770. [Google Scholar] [CrossRef]
- Park, J.A.; Cho, E.J.; Park, J.Y.; Sohn, S.H. Histological change of uterus endometrium and expression of the eggshell-related genes according to hen age. Korean J. Poult. Sci. 2017, 44, 19–28. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Higgs, D.R.; Thomas, R.G. Molecular biology: RNA discrimination. Nature 2012, 482, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Zhou, L.Y.; Xu, S.; Zheng, Y.L.; Wan, Y.F.; Hu, C.P. Overexpression of lncRNA HOXA11-AS promotes cell epithelial–mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Tang, Y.A.; Lu, Y.H.; Lai, W.W.; Wang, Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Teranishi, M.; Shimada, Y.; Hori, T.; Nakabayashi, O.; Kikuchi, T.; Macleod, T.; Pym, R.; Sheldon, B.; Solovei, I.; Macgregor, H.; et al. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res. 2001, 9, 147–165. [Google Scholar] [CrossRef]
- Roeszler, K.N.; Itman, C.; Sinclair, A.H.; Smith, C.A. The long non-coding RNA, MHM, plays a role in chicken embryonic development, including gonadogenesis. Dev. Biol. 2012, 366, 317–326. [Google Scholar] [CrossRef]
- Arriaga-Canon, C.; Fonseca-Guzmán, Y.; Valdes-Quezada, C.; Arzate-Mejía, R.; Guerrero, G.; Recillas-Targa, F. A long non-coding RNA promotes full activation of adult gene expression in the chicken α-globin domain. Epigenetics 2014, 9, 173–181. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhang, K.; Shi, Z.; Zhang, J.; Zhu, J.; Zhu, S.; Zhang, A.; Jia, Z.; Wang, G.; Yu, S.; et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012, 40, 2004. [Google Scholar] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Canessa, C.M.; Schild, L.; Buell, G.; Thorens, B.; Gautschi, I.; Horisberger, J.D.; Rossier, B.C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 1994, 367, 463–467. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, H.; Li, Z.; Zheng, X.; Jia, X.; Nie, Q.; Zhang, X. Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers. PLoS ONE 2013, 8, e56411. [Google Scholar] [CrossRef]
- Adiconis, X.; Borges-Rivera, D.; Satija, R.; DeLuca, D.S.; Busby, M.A.; Berlin, A.M.; Sivachenko, A.; Thompson, D.A.; Wysoker, A.; Fennell, T.; et al. Corrigendum: Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods 2013, 10, 623. [Google Scholar] [CrossRef]
- Hung, T.; Chang, H.Y. Long noncoding RNA in genome regulation. RNA Biol. 2010, 7, 582–585. [Google Scholar] [CrossRef]
- Dong, R.; Jia, D.; Xue, P.; Cui, X.; Li, K.; Zheng, S.; He, X.; Dong, K. Genome-Wide Analysis of Long Noncoding RNA (lncRNA) Expression in Hepatoblastoma Tissues. PLoS ONE 2014, 9, e85599. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, X.; Yang, W.; Lu, Z.; Wang, P.; Chen, Z.; He, J. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumor Biol. 2016, 37, 13091–13100. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front. Physiol. 2017, 7, 687. [Google Scholar] [CrossRef]
- Jonchère, V.; Réhault-Godbert, S.; Hennequet-Antier, C.; Cabau, C.; Sibut, V.; Cogburn, L.A.; Nys, Y.; Gautron, J. Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC Genom. 2010, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Jonchère, V.; Brionne, A.; Gautron, J.; Nys, Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 2012, 12, 10. [Google Scholar] [CrossRef]
- Brionne, A.; Nys, Y.; Hennequet-Antier, C.; Gautron, J. Hen uterine gene expression profiling during eggshell formation reveals putative proteins involved in the supply of minerals or in the shell mineralization process. BMC Genom. 2014, 15, 220. [Google Scholar] [CrossRef]
- Du, J.; Hincke, M.T.; Rose-Martel, M.; Hennequet-Antier, C.; Brionne, A.; Cogburn, L.A.; Nys, Y.; Gautron, J. Identifying specific proteins involved in eggshell membrane formation using gene expression analysis and bioinformatics. BMC Genom. 2015, 16, 792. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, F.; Liu, L.; Zheng, C.W.; Wang, D.H.; Hou, Z.C.; Ning, Z.H. Integrating transcriptome and genome re-sequencing data to identify key genes and mutations affecting chicken eggshell qualities. PLoS ONE 2015, 10, e0125890. [Google Scholar] [CrossRef]
- Jung, J.G.; Lim, W.; Park, T.S.; Kim, J.N.; Han, B.K.; Song, G.; Han, J.Y. Structural and histological characterization of oviductal magnum and lectin-binding patterns in Gallus domesticus. Reprod. Biol. Endocrinol. 2011, 9, 62. [Google Scholar] [CrossRef]
- Nys, Y.; Gautron, J.; Garcia-Ruiz, J.M.; Hinckec, M.T. Avian eggshell mineralization: Biochemical and functional characterization of matrix proteins. Comptes Rendus Palevol 2004, 3, 549–562. [Google Scholar] [CrossRef]
- Arad, Z.; Eylath, U.; Ginsburg, M.; Eyal-Giladi, H. Changes in uterine fluid composition and acid-base status during shell formation in the chicken. Am. J. Physiol. 1989, 257, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Panhéleux, M.; Kälin, O.; Gautron, J.; Nys, Y. Features of eggshell formation in guinea fowl: Kinetics of shell deposition, uterine protein secretion and uterine histology. Br. Poult. Sci. 1999, 40, 632. [Google Scholar] [CrossRef]
- Nakada, T.; Koga, O. Stimulation of secretion of shell gland fluid and calcium by the presence of ovum or ovum-like mass containing artificial yolk in the oviduct uterus of the hen. J. Poult. Sci. 2008, 27, 21–28. [Google Scholar] [CrossRef]
- Benos, D.J.; Stanton, B.A. Functional domains within the degenerin/epithelial sodium channel (Deg/ENaC) superfamily of ion channels. J. Physiol. 1999, 520, 631–644. [Google Scholar] [CrossRef]
- Parker, S.L.; Lindsay, L.A.; Herbert, J.F.; Murphy, C.R.; Thompson, M.B. Expression and localization of Ca2+-ATPase in the uterus during the reproductive cycle of king quail (Coturnix chinensis) and zebra finch (Poephila guttata). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2008, 149, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R. The phosphatonins and the regulation of phosphate homeostasis. Am. J. Physiol. Renal. Physiol. 2006, 67, 142–146. [Google Scholar] [CrossRef]
- Foster, B.L.; Tompkins, K.A.; Rutherford, R.B.; Zhang, H.; Chu, E.Y.; Fong, H.; Somerman, M.J. Phosphate: Known and potential roles during development and regeneration of teeth and supporting structures. Birth Defects Res. Part C Embryo Today Rev. 2008, 84, 281–314. [Google Scholar] [CrossRef]
- Yoshioka, H.; Yoshiko, Y.; Minamizaki, T.; Suzuki, S.; Koma, Y.; Nobukiyo, A.; Sotomaru, Y.; Suzuki, A.; Itoh, M.; Maeda, N. Incisor enamel formation is impaired in transgenic rats overexpressing the type III NaPi transporter Slc20a1. Calcif. Tissue Int. 2011, 89, 192–202. [Google Scholar] [CrossRef]
- Itoh, N.; Ornitz, D.M. Functional evolutionary history of the mouse Fgf gene family. Dev. Dyn. 2008, 237, 18–27. [Google Scholar] [CrossRef]
- Ali, S.; Shavkunov, A.; Panova, N.; Stoilova-McPhie, S.; Laezza, F. Modulation of the FGF14:FGF14 homodimer interaction through short peptide fragments. CNS Neurol. Disord. Drug Targets 2014, 13, 1559–1570. [Google Scholar] [CrossRef]
- Yan, H.; Pablo, J.L.; Pitt, G.S. FGF14 regulates presynaptic Ca2+ channels and synaptic transmission. Cell Rep. 2013, 11, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Xu, Y.; Scearce-Levie, K.; Ptácek, L.J.; Fu, Y.H. COL25A1 triggers and promotes Alzheimer’s disease-like pathology in vivo. Neurogenetics 2010, 11, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Avenarius, M.R. The Glutaredoxin-Like Cysteine-Rich Family of Genes, Grxcr1 and Grxcr2, in Stereocilia Development and Function. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 2012. [Google Scholar]
- Odeh, H.; Hunker, K.L.; Belyantseva, I.A.; Azaiez, H.; Avenarius, M.R.; Zheng, L.; Peters, L.M.; Gagnon, L.H.; Hagiwara, N.; Skynner, M.J.; et al. Mutations in Grxcr1 are the Basis for Inner Ear Dysfunction in the Pirouette Mouse. Am. J. Hum. Genet. 2010, 86, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Pastorekova, S.; Parkkila, S.; Zavada, J. Tumor-associated Carbonic Anhydrases and Their Clinical Significance. Adv. Clin. Chem. 2006, 42, 167. [Google Scholar] [PubMed]
- Pines, M.; Knopov, V.; Bar, A. Involvement of osteopontin in eggshell formation in the laying chicken. Matrix Biol. J. Int. Soc. Matrix Biol. 1995, 14, 765. [Google Scholar] [CrossRef]
- Huang, Z.; Rose, A.H.; Hoffmann, P.R. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2012, 16, 705–743. [Google Scholar] [CrossRef]
- Yoboue, E.D.; Rimessi, A.; Anelli, T.; Pinton, P.; Sitia, R. Regulation of Calcium Fluxes by GPX8, a Type-II Transmembrane Peroxidase Enriched at the Mitochondria-Associated Endoplasmic Reticulum Membrane. Antioxid. Redox Signal. 2017, 27, 583–595. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).