Abstract
Chromosome 4p deletions can lead to two distinct phenotypic outcomes: Wolf-–Hirschhorn syndrome (a terminal deletion at 4p16.3) and less frequently reported proximal interstitial deletions (4p11-p16). Proximal 4p interstitial deletions can result in mild to moderate intellectual disability, facial dysmorphisms, and a tall thin body habitus. To date, only 35 cases of proximal 4p interstitial deletions have been reported, and only two of these cases have been familial. The critical region for this syndrome has been narrowed down to 4p15.33-15.2, but the underlying causative genes remain unclear. In this study, we report the case of a 3-year-old female with failure to thrive, developmental and motor delays, and morphological features. The mother also had a 4p15.2-p14 deletion, and the proband was found to have a 13.4-Mb 4p15.2-p14 deletion by chromosome microarray analysis. The deleted region encompasses 16 genes, five of which have a high likelihood of contributing to the phenotype: PPARGC1A, DHX15, RBPJ, STIM2, and PCDH7. These findings suggest that multiple genes are involved in this rare proximal 4p interstitial deletion syndrome. This case highlights the need for healthcare providers to be aware of proximal 4p interstitial deletions and the potential phenotypic manifestations.
1. Introduction
Two distinct phenotypes have been associated with deletions on chromosome 4p: Wolf-Hirschhorn syndrome (WHS; OMIM 194190)—terminal deletion [1], which is characterized by significant growth and severe intellectual disability, seizures, specific facial deformities (Greek warrior helmet appearance), and multiple congenital anomalies; and proximal interstitial deletions (4p11-p16) which have been rarely reported and may have distinct mild to moderate intellectual disability, multiple dysmorphic features including long face, up-slanted palpebral fissures with epicanthal folds, tall thin body habitus, and hyperextensible joints [2,3,4,5,6,7,8,9,10,11,12,13,14,15]. The phenotypic manifestation of proximal 4p deletion syndrome is generally less severe than that of WHS but is not well-known among healthcare professionals and confers a broad spectrum of congenital abnormalities. It may thus go undiagnosed until the affected person is older. Upon literature review, 35 cases with interstitial deletions of 4p have been described, with ages from prenatal to 77-year-old (mean 15.1, median 30), while males and females were equally distributed. To date, only two familial cases have been reported, and most of the reported deletions were detected by conventional chromosome analyses [3,7]. In addition, the critical region for proximal 4p interstitial deletion syndrome has been localized to 4p15.33-15.2, but the causative gene or genes remain elusive. In this study, we report a rare familial proximal interstitial 4p deletion case with an assessment of the pertinent deleted genes and a thorough literature review of similar deletions. We hope this report will help make this syndrome more recognizable among clinicians and provide a foundation for identifying the primary causative gene (or genes).
2. Case Presentation
A 3-year-old female, referred to as the proband, was brought to the Clinical Genetic Laboratories at the University of Kentucky for examination and testing. She was experiencing growth failure, delays in her physical and intellectual development, and had physical features that were of unknown origin. The proband appeared thin, had a small head with normal hair, larger ears, long eyelashes, upward-slanting eye openings, a low nasal bridge with broad nostrils, and a heart-shaped upper lip with a grooved philtrum, and a slightly enlarged tongue.
The proband’s mother, at the age of 23, had an intellectual disability and physical abnormalities including large knuckles, a tall and slender physique, a heart murmur, 50% loss of hearing in the left ear, vision abnormalities, and facial abnormalities. Her karyotype was previously tested when she was 9 years old and showed a deletion in the 4p15.2-p14 region. She was evaluated again at 10 years and 10 months and was found to have borderline microcephaly, anxiety, and frequent laughing, a flat mid-face, upward-slanting eye openings, folds over the inner corners of her eyes, borderline wide-set eyes, an upturned nasal tip, a narrow palate, a right crease, crowded teeth, bifid uvula, hypermobility of elbows, and developmental delays. She was born full-term and without any complications, weighing 6 lb 5 oz and measuring 19 inches long. Genetic testing was performed due to her distinctive physical features.
The proband also had two maternal uncles, ages 16 and 12, who were healthy and two maternal aunts, ages 19 and 5, who were also healthy, but their karyotypes were unavailable. The karyotype of the maternal grandmother was reported as normal. Multiple family members on her maternal grandfather’s side are said to have a similar appearance to the proband’s mother and her. The proband’s father was 29 years old and healthy. Both parents are Caucasian and are not known to be consanguineous.
3. Methods and Results
3.1. Cytogenetic Analysis
Using standard procedures, routine G-banded karyotyping was performed on cultured peripheral blood lymphocytes from the patient and her mother, at the 550-band level according to the International System for Human Cytogenomic Nomenclature (ISCN) 2020. Conventional chromosome studies on this proband showed a deletion of 4p15.2-p14 (Figure 1), the same as her mother.
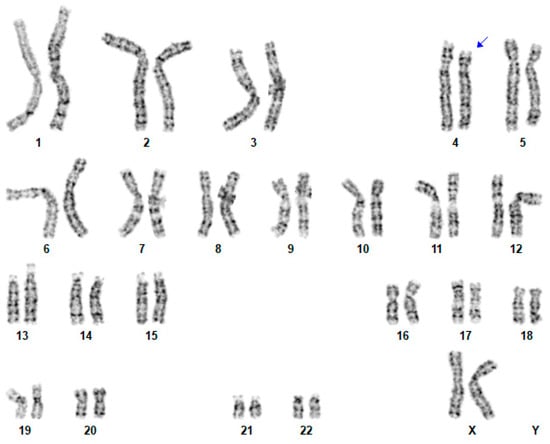
Figure 1.
Conventional cytogenetic analysis on this proband. A karyotype 46,XX,del(4)(p15.2-p14). Blue arrow indicates where the proximal deletion is located.
3.2. High-Resolution SNP Array Analysis
Genomic DNA was isolated from peripheral blood lymphocytes with the QIAamp DNA Blood Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer’s instructions. DNA sample (250 ng) of the proband was hybridized to CytoScan HD arrays on an Affymetrix SNP array platform (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The CytoScan HD array contains more than 2.6 million markers for the copy number analysis. Of these markers, 1,950,000 are unique, non-polymorphic oligonucleotide probes, and 750,000 are SNP probes used for genotyping. The average marker spacing is one probe per 1.1 kb, with a mean spacing of one probe per 1.7 kb on non-gene backbones and one probe per 880 bp in intragenic regions. The ChAS 4.2.0.80 software (Affymetrix; Thermo Fisher Scientific, Inc) was used for copy number variants analyses. A 13.4-Mb deletion at 4p15.2-p14 encompassing 16 genes was identified on the proband (Table 1; Figure 2).

Table 1.
Genes deleted in the 4p15.2-14 region in the presented case.
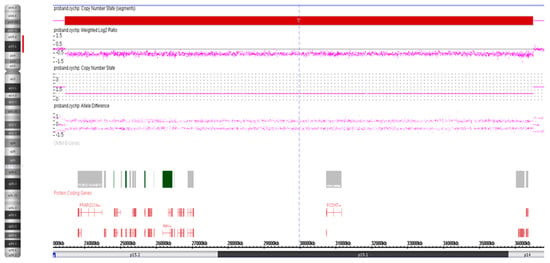
Figure 2.
Whole-genome CMA analysis on uncultured peripheral blood lymphocytes of the proband. An 13.4-Mb deletion at 4p15.2-p14 [ISCN arr[GRCh37] 4p15.2-p14(23443552_36483601)×1].
The clinical interpretation of this deletion was based on a scoring system recommended by the ACMG (American College of Medical Genetics) guidelines published in 2019 [16]. The available evidence in the literature is not enough to classify this finding as a pathogenic variant. However, the relatively consistent clinical manifestation pattern in many of the 35 previous cases (plus the two described here), and the similar genomic locations on 4p, suggest the possibility that this may be a recognizable genetic syndrome. Our molecular analyses show 16 genes in this deleted region for our proband, and five of these 16 genes, PPARGC1A, DHX15, RBPJ, STIM2 and PCDH7, have a pLI score of more than 0.98, and thus likely to have phenotypic effects. These molecular findings combined with the prior clinical and cytogenetic studies may strengthen the case for defining a new proximal 4p interstitial deletion syndrome that is distinct from the well-established 4p16.3 (WHS) terminal deletion syndrome.
4. Discussion
Proximal interstitial deletions of chromosome 4p are relatively rare. Upon careful literature review, fewer than 40 patients have been described (Table S1). The age range is from prenatal to 77-year-old (mean 15.1, median 30), and females and males are equally distributed. The main clinical features of the reported cases including the presented cases are mild to moderate intellectual disability (96%), while over half the patients have multiple minor dysmorphic features including a long face, midface hypoplasia, upslanted fissures, epicanthal folds, large beaked nose, thick lower lip, high palate, and tall-thin body habitus. In addition, more than half of male patients have cryptorchidism and about 40% of patients show congenital heart diseases, including pulmonary stenosis, atrial septal defect, and Tetralogy of Fallot. Compared to “significant growth and severe intellectual disability, seizures, specific facial deformities (Greek warrior helmet appearance)” [1], manifestations of WHS, the phenotype of proximal 4p interstitial deletion syndrome is usually relatively mild and grants a broad spectrum of congenital abnormalities, however, it is not well recognized among health care providers. Therefore, it may be overlooked till the patient is older [17].
The critical region for proximal 4p deletion syndrome has been localized to 4p15.33-15.2, which is also distinct from the terminal location of WHS. Since most studies used traditional cytogenetic analysis, the causative gene or genes remain unknown. In this study, we found 16 genes in the deletion region, five of which are more likely to have phenotypic effects due to haploinsufficiency (pLI scores > 0.98). One or more of these five genes are likely to be the primary causative factors in the clinical manifestations. To our knowledge, there have been only two reported instances of familial interstitial 4p deletion, both of which were identified by conventional chromosome analyses [3,7]. A more detailed illustration of the genes implicated in the deleted region would improve our understanding of the correlation between genotype and phenotype. Recent reports have identified de novo interstitial 4p deletions using CMA and conventional G-banding cytogenetics similar to the methods used in our study. Mitroi et al. [13] and Park et al. [17] identified de novo 4p interstitial deletions in a proband with intellectual disability, mild dysmorphic features, and mild hypotonia, similar to our case. The authors speculated that SLIT2, KCNIP4, RBPJ, and LG12 genes are involved in neurologic development and skeletal abnormalities. The genes identified in our study significantly overlap with previous studies, however, we did not identify SLIT2 and KCNIP4 genes in the deleted 4p region. The variation in the methodology employed across different studies may, to some extent, account for the discrepancy.
Recombination signal-binding protein for immunoglobulin Kappa J region (RBPJ) encoded by RBPJ gene, is a transcriptional regulator playing a critical role in the Notch signaling pathway [18]. As described by Giaimo et al. [19], RBPJ can behave as a “molecular switch” from activation to repression Notch signal transduction cascades during blood cell development. The on/off switch of Notch signaling is well known to regulate cell proliferation and apoptosis in multiple organs [20]. Previous studies have also shown that the reduced expression of Notch pathway proteins can lead to multiple developmental defects [21,22], suggesting that Notch pathway is likely dosage dependent. Haploinsufficiency of Notch signaling components including RBPJ gene has been reported in families with Adams-Oliver syndrome (AOS [MIM 100300]) [23], which usually shows autosomal-dominant inheritance, with the most common features being terminal limb malformations and congenital cutis aplasia [23]. The presented proband has haploinsufficiency of RBPJ and shows mainly facial deformations, long/thin body status, and her mother had congenital cardiac disease but didn’t show severe vascular and limb defects of AOS, therefore, other components of the pathway must have played a role. For example, RBPJ-mediated Notch signaling strength has been shown to be involved in mesenchymal cell proliferation and skeletal formation [24], epidermis and hair follicle development [25], and vascular structure formation [26]. Moreover, Krebs et al. showed that RBPJ conditional-knockout mice have arteriovenous malformations [18]. The disruption of RBPJ by a translocation was also reported in an individual who presented with a similar phenotype to proximal 4p deletion [27].
The stromal interaction molecule STIM1 and STIM2 function as sensors of Ca2+ in the endoplasmic reticulum and maintain high cytosolic calcium levels by regulating Ca2+ influx [28]. Altered expressions of STIM1 in mouse and cell models have demonstrated its critical function in hypertension, adipocyte differentiation, cognitive impairment, and cancer [29,30,31]. Unlike STIM1, the function of STIM2 has not been fully explored. STIM1-deficient mice have been shown perinatally lethal, while STIM2-deficient mice show a growth delay and begin to die around 4–8 weeks after birth [31,32,33]. STIM2 seems to protect hippocampal neurons from amyloid synaptotoxicity [34], and inhibition of STIM2 by miR-128 causing memory impairment in Alzheimer’s disease mouse model [35], which suggests the neuroprotective potentials of STIM2 [36]. Recent studies also show that STIM2 involves the invasion and metastasis of breast cancer [37] and cancer-induced inflammation in leukemia [38]. The DEAH-box RNA helicase 15 (DHX15) is implicated in RNA metabolism, including splicing and ribosome biogenesis [39,40]. It’s also involved in several biological processes, such as regulating antiviral innate immune responses in enterocytes [41] which is recently revealed crystal structure by Murakami et al. [42]. Many studies have discussed the involvement of DHX15 in several cancer progression, either enhancing or suppressing it [43,44,45]. More importantly, DHX15 gene knockdown in Jurkat, raji and NB4 cells by Chen et al. and Pan et al. groups have shown to induce cell apoptosis, arrest cell cycle, and inhibit cell proliferation [43,46]. The haploinsufficiency of DHX15 in our proband could imply the involvement of DHX15 in the facial dysmorphic features and developmental delay.
Peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1α/PPARGC1A) is a crucial transcriptional coactivator required for lipid metabolism in the mitochondria [47], implicated in the pathogenesis of neurodegenerative disorders [48]. In addition, the role of PGC-1α in the pathogenesis of age-related macular degeneration is related to direct or indirect interactions with vascular endothelial growth factor (VEGF), telomerase, oxidative stress, and autophagy-related mechanisms [49]. PGC-1α has unique roles in the central nervous system of mouse models, such as in the maturation of neuronal cells and synaptogenesis, while deficiencies promote behavioral dysfunction by impacting the inhibitory signals of dopaminergic and interneurons [50]. PCDH7 encodes the cadherin protein of brain–heart protocadherin (BHPCDH), which mediates calcium-dependent cell-cell adhesion [51]. PCDH genes are essential for brain development [52]. PCDH7 neural fold protocadherin (NFPC) has been shown to regulate differentiation of the embryonic ectoderm [53], neural tube formation [54], and cell morphology [55]. PCDH7 promoter activities have been observed to be down-regulated by MECP2, and mutations of the MECP2 gene cause MECP2 reduction and BHPCDH up-regulation in postmortem brains from Rett syndrome patients [56]. These findings suggest that haploinsufficiency of PCDH7 may be involved in the mental developmental delay of our proband and likely other proximal 4p deletion syndrome patients. However, the understanding of the molecular mechanisms of PCDH7 is still poor and requires further study.
5. Conclusions
We reviewed the phenotypic abnormalities of previously reported patients with similar proximal 4p interstitial deletions and discussed the gene functions deleted within this region. This report contributes to expanded knowledge of the genetic background of proximal 4p interstitial deletion syndrome and facilitates accurate genetic counseling, including recurrence risks and prenatal testing options. The genetic investigations performed in the study have allowed us to define better the critical chromosomal region and the best candidate genes for follow-up studies. We anticipate this is an emerging distinct genetic syndrome that is proximal to the well-described 4p16.3 (WHS) terminal deletion syndrome.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14030635/s1, Table S1: Phenotypic features of reported interstitial 4p deletion cases during the past 45 years.
Author Contributions
J.D. and L.Y. contributed to the manuscript writing and analyses, H.T.R. contributed to the patient phenotype and family history. S.Z. and S.W. provided laboratory technical guidance and interpretation. S.W. initiated and oversaw the research effort, writing, and other aspects of this project. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The Institutional Review Board at the University of Kentucky Health Center had granted an exemption for ethical approval, and informed consent was obtained from the proband’s legal guardians in accordance with the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper. Ethical code: Medical research.
Data Availability Statement
All data that supports the findings of this study are included in this published article and its supplementary information files.
Acknowledgments
The authors would like to express their gratitude to the family of the proband for their participation in this study. We also acknowledge the support of the Clinical Genetic Laboratories at the University of Kentucky for their expertise in performing the conventional chromosome and CMA studies. The authors would also like to extend their thanks to the Institutional Review Board at the University of Kentucky Health Center for granting ethical approval for this study. Finally, the authors would like to acknowledge the support of their colleagues and the contribution of their research team in making this study a success.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Altherr, M.R.; Bengtsson, U.; Elder, F.F.; Ledbetter, D.H.; Wasmuth, J.J.; McDonald, M.E.; Gusella, J.F.; Greenberg, F. Molecular confirmation of Wolf-Hirschhorn syndrome with a subtle translocation of chromosome 4. Am. J. Hum. Genet. 1991, 49, 1235–1242. [Google Scholar]
- Alesi, V.; Barrano, G.; Morara, S.; Darelli, D.; Petrilli, K.; Capalbo, A.; Pacella, M.; Haass, C.; Finocchi, M.; Novelli, A.; et al. A previously undescribed de novo 4p15 deletion in a patient with apparently isolated metopic craniosynostosis. Am. J. Med. Genet. A 2011, 155A, 2543–2551. [Google Scholar] [CrossRef]
- Basinko, A.; Douet-Guilbert, N.; Parent, P.; Blondin, G.; Mingam, M.; Monot, F.; Morel, F.; Le Bris, M.-J.; De Braekeleer, M. Familial interstitial deletion of the short arm of chromosome 4 (p15.33–p16.3) characterized by molecular cytogenetic analysis. Am. J. Med. Genet. Part A 2008, 146A, 899–903. [Google Scholar] [CrossRef]
- Estabrooks, L.L.; Rao, K.W.; Korf, B. Interstitial deletion of distal chromosome 4p in a patient without classical Wolf-Hirschhorn syndrome. Am. J. Med. Genet. 1993, 45, 97–100. [Google Scholar] [CrossRef] [PubMed]
- White, D.M.; Pillers, D.-A.M.; Reiss, J.A.; Brown, M.G.; Magenis, R.E. Interstitial deletions of the short arm of chromosome 4 in patients with a similar combination of multiple minor anomalies and mental retardation. Am. J. Med. Genet. 1995, 57, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Van De Graaf, G.; Sijstermans, J.M.; Engelen, J.J.; Schrander-Stumpel, C.T. Mild phenotype in interstitial 4p deletion: Another patient and review of the literature. Genet. Couns. 1997, 8, 13–18. [Google Scholar] [PubMed]
- Tonk, V.S.; Jalal, S.M.; Gonzalez, J.; Kennedy, A.; Velagaleti, G.V. Familial interstitial deletion of chromosome 4 (p15.2p16.1). Ann. Genet. 2003, 46, 453–458. [Google Scholar] [CrossRef]
- Su, P.-H.; Lee, I.-C.; Chen, J.-Y.; Chen, S.-J.; Yu, J.-S.; Tsao, T.-F. Interstitial Deletions of the Short Arm of Chromosome 4 in a Patient With Mental Retardation and Focal Seizure. Pediatr. Neonatol. 2011, 52, 165–168. [Google Scholar] [CrossRef]
- South, S.T.; Corson, V.L.; McMichael, J.L.; Blakemore, K.J.; Stetten, G. Prenatal Detection of an Interstitial Deletion in 4p15 in a Fetus with an Increased Nuchal Skin Fold Measurement. Fetal Diagn. Ther. 2004, 20, 58–63. [Google Scholar] [CrossRef]
- Ray, M.; Evans, J.; Rockman-Greenberg, C.; Wickstrom, D. Interstitial deletion of the short arm of chromosome 4. J. Med. Genet. 1984, 21, 223–225. [Google Scholar] [CrossRef]
- Piovani, G.; Borsani, G.; Bertini, V.; Kalscheuer, V.M.; Viertel, P.; Bellotti, D.; Valseriati, D.; Barlati, S. Unexpected identification of two interstitial deletions in a patient with a pericentric inversion of a chromosome 4 and an abnormal phenotype. Eur. J. Med. Genet. 2006, 49, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Møller, R.; Hansen, C.; Ullmann, R.; Ropers, H.; Tommerup, N.; Tümer, Z.; Jackson, G. Interstitial deletion of chromosome 4p associated with mild mental retardation, epilepsy and polymicrogyria of the left temporal lobe. Clin. Genet. 2007, 72, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Mitroi, A.; Aschie, M.; Apostol, A.; Brinzan, C.; Cozaru, G.; Mitroi, A.N. A boy with 13.34-Mb interstitial deletion of chromosome 4p15: A new case report and review of the literature. Medicine 2017, 96, e9301. [Google Scholar] [CrossRef]
- Ishikawa, T.; Sumi, S.; Fujimoto, S.; Shima, Y.; Wada, Y. Interstitial deletion of the short arm of chromosome 4 in a boy with mild psychomotor retardation and dysmorphism. Clin. Genet. 1990, 38, 314–317. [Google Scholar] [CrossRef]
- Fryns, J.P.; Yang-Aisheng; Kleczkowska, A.; Lemmens, F.; Vandecasseye, W.; van den Berghe, H. Interstitial deletion of the short arm of chromosome 4. A phenotype distinct from the Wolf-Hirschhorn syndrome. Ann. Genet. 1989, 32, 59–61. [Google Scholar] [PubMed]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jeon, B.R.; Lee, Y.K.; Ki, C.-S.; Jang, M.-A. The First Korean Case of De Novo Proximal 4p Deletion Syndrome in a Child With Developmental Delay. Ann. Lab. Med. 2020, 40, 435–437. [Google Scholar] [CrossRef] [PubMed]
- Krebs, L.T.; Shutter, J.R.; Tanigaki, K.; Honjo, T.; Stark, K.L.; Gridley, T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004, 18, 2469–2473. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Gagliani, E.K.; Kovall, R.A.; Borggrefe, T. Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response. Adv. Exp. Med. Biol. 2020, 1287, 9–30. [Google Scholar] [CrossRef]
- Shen, W.; Huang, J.; Wang, Y. Biological Significance of NOTCH Signaling Strength. Front. Cell Dev. Biol. 2021, 9, 652273. [Google Scholar] [CrossRef]
- Blackwood, C.A.; Bailetti, A.; Nandi, S.; Gridley, T.; Hébert, J.M. Notch Dosage: Jagged1 Haploinsufficiency Is Associated With Reduced Neuronal Division and Disruption of Periglomerular Interneurons in Mice. Front. Cell Dev. Biol. 2020, 8, 113. [Google Scholar] [CrossRef]
- McCright, B.; Lozier, J.; Gridley, T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 2002, 129, 1075–1082. [Google Scholar] [CrossRef]
- Hassed, S.J.; Wiley, G.B.; Wang, S.; Lee, J.-Y.; Li, S.; Xu, W.; Zhao, Z.J.; Mulvihill, J.J.; Robertson, J.; Warner, J.; et al. RBPJ Mutations Identified in Two Families Affected by Adams-Oliver Syndrome. Am. J. Hum. Genet. 2012, 91, 391–395. [Google Scholar] [CrossRef]
- Dong, Y.; Jesse, A.M.; Kohn, A.; Gunnell, L.M.; Honjo, T.; Zuscik, M.; O'Keefe, R.J.; Hilton, M.J. RBPjκ-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 2010, 137, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Vauclair, S.; Nicolas, M.; Barrandon, Y.; Radtke, F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol. 2005, 284, 184–193. [Google Scholar] [CrossRef]
- Dou, G.; Wang, Y.; Hu, X.; Hou, L.; Wang, C.; Xu, J.; Wang, Y.; Liang, Y.; Yao, L.; Yang, A.; et al. RBP-J, the transcription factor downstream of Notch receptors, is essential for the maintenance of vascular homeostasis in adult mice. FASEB J. 2007, 22, 1606–1617. [Google Scholar] [CrossRef]
- Nakayama, T.; Saitsu, H.; Endo, W.; Kikuchi, A.; Uematsu, M.; Haginoya, K.; Hino-Fukuyo, N.; Kobayashi, T.; Iwasaki, M.; Tominaga, T.; et al. RBPJ is disrupted in a case of proximal 4p deletion syndrome with epilepsy. Brain Dev. 2013, 36, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Soboloff, J.; Spassova, M.A.; Tang, X.D.; Hewavitharana, T.; Xu, W.; Gill, D.L. Orai1 and STIM Reconstitute Store-operated Calcium Channel Function. J. Biol. Chem. 2006, 281, 20661–20665. [Google Scholar] [CrossRef] [PubMed]
- Mancarella, S.; Potireddy, S.; Wang, Y.; Gao, H.; Gandhirajan, R.K.; Autieri, M.; Scalia, R.; Cheng, Z.; Wang, H.; Madesh, M.; et al. Targeted STIM deletion impairs calcium homeostasis, NFAT activation, and growth of smooth muscle. FASEB J. 2012, 27, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.T.; Alevizos, I.; Liu, X.; Swaim, W.D.; Yin, H.; Feske, S.; Oh-Hora, M.; Ambudkar, I.S. STIM1 and STIM2 protein deficiency in T lymphocytes underlies development of the exocrine gland autoimmune disease, Sjögren’s syndrome. Proc. Natl. Acad. Sci. USA 2012, 109, 14544–14549. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, L.S.; Graham, S.J.L.; Dziadek, M.A. STIM proteins: Integrators of signalling pathways in development, differentiation and disease. J. Cell. Mol. Med. 2010, 14, 1890–1903. [Google Scholar] [CrossRef] [PubMed]
- Picard, C.; McCarl, C.-A.; Papolos, A.; Khalil, S.; Lüthy, K.; Hivroz, C.; LeDeist, F.; Rieux-Laucat, F.; Rechavi, G.; Rao, A.; et al. STIM1 Mutation Associated with a Syndrome of Immunodeficiency and Autoimmunity. N. Engl. J. Med. 2009, 360, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Oh-Hora, M.; Yamashita, M.; Hogan, P.G.; Sharma, S.; Lamperti, E.; Chung, W.; Prakriya, M.; Feske, S.; Rao, A. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 2008, 9, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Popugaeva, E.; Pchitskaya, E.; Speshilova, A.; Alexandrov, S.; Zhang, H.; Vlasova, O.; Bezprozvanny, I. STIM2 protects hippocampal mushroom spines from amyloid synaptotoxicity. Mol. Neurodegener. 2015, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Zhang, Q.; Wu, Z.; Ma, T.; He, A.; Zhang, T.; Ke, X.; Yu, Q.; Han, Y.; Lu, Y. Mossy cell synaptic dysfunction causes memory imprecision via miR-128 inhibition of STIM2 in Alzheimer's disease mouse model. Aging Cell 2020, 19, e13144. [Google Scholar] [CrossRef]
- Serwach, K.; Gruszczynska-Biegala, J. Target Molecules of STIM Proteins in the Central Nervous System. Front. Mol. Neurosci. 2020, 13, 617422. [Google Scholar] [CrossRef]
- Cheng, H.; Chen, L.; Fang, Z.; Wan, Q.; Du, Z.; Ma, N.; Guo, G.; Lu, W. STIM2 promotes the invasion and metastasis of breast cancer cells through the NFAT1/TGF-β1 pathway. Cell. Mol. Biol. 2022, 67, 55–61. [Google Scholar] [CrossRef]
- Fleur-Lominy, S.S.; Maus, M.; Vaeth, M.; Lange, I.; Zee, I.; Suh, D.; Liu, C.; Wu, X.; Tikhonova, A.; Aifantis, I.; et al. STIM1 and STIM2 Mediate Cancer-Induced Inflammation in T Cell Acute Lymphoblastic Leukemia. Cell Rep. 2018, 24, 3045–3060.e5. [Google Scholar] [CrossRef]
- Abdelhaleem, M.; Maltais, L.; Wain, H. The human DDX and DHX gene families of putative RNA helicases. Genomics 2003, 81, 618–622. [Google Scholar] [CrossRef]
- Imamura, O.; Sugawara, M.; Furuichi, Y. Cloning and Characterization of a Putative Human RNA Helicase Gene of the DEAH-Box Protein Family. Biochem. Biophys. Res. Commun. 1997, 240, 335–340. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, S.; Yang, L.; Cui, S.; Pan, W.; Jackson, R.; Zheng, Y.; Rongvaux, A.; Sun, Q.; Yang, G.; et al. Nlrp6 regulates intestinal antiviral innate immunity. Science 2015, 350, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Nakano, K.; Shimizu, T.; Ohto, U. The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2017, 73 Pt 6, 347–355. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Pan, L.; Huang, Y.; Cai, Y.; Li, J.; Li, Y.; Wang, S. RNA helicase DHX15 decreases cell apoptosis by NF-κB signaling pathway in Burkitt lymphoma. Cancer Cell Int. 2022, 22, 92. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Chen, K.; Qin, Y.; Niu, Y.; Zhang, X.; Xu, S.; Zhang, C.; Feng, M.; Wang, K. Long Non-coding RNA JHDM1D-AS1 Interacts with DHX15 Protein to Enhance Non-Small-Cell Lung Cancer Growth and Metastasis. Mol. Ther. Nucleic Acids 2019, 18, 831–840. [Google Scholar] [CrossRef]
- Faber, Z.J.; Chen, X.; Gedman, A.L.; Boggs, K.; Cheng, J.; Ma, J.; Radtke, I.; Chao, J.-R.; Walsh, M.P.; Song, G.; et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 2016, 48, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, Y.; Zhang, H.-Y.; Zheng, Y.; Liu, X.-L.; Hu, Z.; Wang, Y.; Wang, J.; Cai, Y.-H.; Liu, Q.; et al. DHX15 is associated with poor prognosis in acute myeloid leukemia (AML) and regulates cell apoptosis via the NF-kB signaling pathway. Oncotarget 2017, 8, 89643–89654. [Google Scholar] [CrossRef]
- Liang, H.; Ward, W.F. PGC-1alpha: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Austin, S.; St-Pierre, J. PGC1α and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012, 125, 4963–4971. [Google Scholar] [CrossRef]
- Blasiak, J.; Szczepanska, J.; Fila, M.; Pawlowska, E.; Kaarniranta, K. Potential of Telomerase in Age-Related Macular Degeneration-Involvement of Senescence, DNA Damage Response and Autophagy and a Key Role of PGC-1α. Int. J. Mol. Sci. 2021, 22, 7194. [Google Scholar] [CrossRef]
- Kuczynska, Z.; Metin, E.; Liput, M.; Buzanska, L. Covering the Role of PGC-1α in the Nervous System. Cells 2021, 11, 111. [Google Scholar] [CrossRef]
- Yoshida, K.; Yoshitomo-Nakagawa, K.; Seki, N.; Sasaki, M.; Sugano, S. Cloning, expression analysis, and chromosomal localization of BH-protocadherin (PCDH7), a novel member of the cadherin superfamily. Genomics 1998, 49, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Yasuda, S.; Tanaka, H.; Yamagata, K.; Kim, H. Non-clustered protocadherin. Cell Adhes. Migr. 2011, 5, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Heggem, M.A.; Bradley, R.S. The cytoplasmic domain of Xenopus NF-protocadherin interacts with TAF1/set. Dev. Cell 2003, 4, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Rashid, D.; Newell, K.; Shama, L.; Bradley, R. A requirement for NF-protocadherin and TAF1/Set in cell adhesion and neural tube formation. Dev. Biol. 2006, 291, 170–181. [Google Scholar] [CrossRef]
- Yoshida, K. Fibroblast cell shape and adhesion in vitro is altered by overexpression of the 7a and 7b isoforms of protocadherin 7, but not the 7c isoform. Cell Mol. Biol. Lett. 2003, 8, 735–741. [Google Scholar]
- Miyake, K.; Hirasawa, T.; Soutome, M.; Itoh, M.; Goto, Y.I.; Endoh, K.; Takahashi, K.; Kudo, S.; Nakagawa, T.; Yokoi, S.; et al. The protocadherins, PCDHB1 and PCDH7, are regulated by MeCP2 in neuronal cells and brain tissues: Implication for pathogenesis of Rett syndrome. BMC Neurosci. 2011, 12, 81. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).