Genetic Variants of HOTAIR Associated with Colorectal Cancer: A Case-Control Study in the Saudi Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Subject Requirement
2.2. Sample Collection
2.3. SNP Selection and Genotyping
2.4. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almatroudi, A. The Incidence Rate of Colorectal Cancer in Saudi Arabia: An Observational Descriptive Epidemiological Analysis. Int. J. Gen. Med. 2020, 13, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, A.; Rudzki, G.; Lewandowski, T.; Stryjkowska-Góra, A.; Rudzki, S. Title: Risk Factors for the Diagnosis of Colorectal Cancer. Cancer Control 2022, 29, 10732748211056692. [Google Scholar] [CrossRef] [PubMed]
- Danial, D.; Youssef, E.D.; Maryam, B.M.; Mohammad, A.; Moein, B.M.; Liliane, D. Risk Factors of Young-Onset Colorectal Cancer: Analysis of a Large Population-Based Registry. Can. J. Gastroenterol. Hepatol. 2022, 2022, 3582443. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Du, M.; Wang, K.; Khandpur, N.; Rossato, S.L.; Drouin-Chartier, J.-P.; Steele, E.M.; Giovannucci, E.; Song, M.; Zhang, F.F. Association of ultra-processed food consumption with colorectal cancer risk among men and women: Results from three prospective US cohort studies. BMJ 2022, 378, e068921. [Google Scholar] [CrossRef]
- Kreile, M.; Piekuse, L.; Rots, D.; Dobele, Z.; Kovalova, Z.; Lace, B. Analysis of possible genetic risk factors contributing to development of childhood acute lymphoblastic leukaemiain the Latvian population. Arch. Med. Sci. 2016, 12, 479–485. [Google Scholar] [CrossRef]
- Grotenhuis, A.J.; Dudek, A.M.; Verhaegh, G.W.; Aben, K.K.; Witjes, J.A.; Kiemeney, L.A.; Vermeulen, S.H. Independent Replication of Published Germline Polymorphisms Associated with Urinary Bladder Cancer Prognosis and Treatment Response. Bl. Cancer 2016, 2, 77–89. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, T.; Song, X.; Dong, G.; Xu, L.; Jiang, F. SNP-Target Genes Interaction Perturbing the Cancer Risk in the Post-GWAS. Cancers 2022, 14, 5636. [Google Scholar] [CrossRef]
- Xu, M.; Gao, Y.; Yu, T.; Wang, J.; Cheng, L.; Cheng, D.; Zhu, B. Functional promoter rs2295080 T>G variant in MTOR gene is associated with risk of colorectal cancer in a Chinese population. Biomed. Pharmacother. 2015, 70, 28–32. [Google Scholar] [CrossRef]
- Li, L.; Sun, R.; Liang, Y.; Pan, X.; Li, Z.; Bai, P.; Zeng, X.; Zhang, D.; Zhang, L.; Gao, L. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J. Exp. Clin. Cancer Res. 2013, 32, 104. [Google Scholar] [CrossRef]
- Xiong, H.Y.; Alipanahi, B.; Lee, L.J.; Bretschneider, H.; Merico, D.; Yuen, R.K.C.; Hua, Y.; Gueroussov, S.; Najafabadi, H.S.; Hughes, T.R.; et al. The human splicing code reveals new insights into the genetic determinants of disease. Science 2015, 347, 1254806. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, J.-W.; He, C.-Y.; Sun, L.-P.; Gong, Y.-H.; Jing, J.-J.; Xing, C.-Z.; Yuan, Y. The Interaction Effects of pri-let-7a-1 rs10739971 with PGC and ERCC6 Gene Polymorphisms in Gastric Cancer and Atrophic Gastritis. PLoS ONE 2014, 9, e89203. [Google Scholar] [CrossRef]
- Janaththani, P.; Srinivasan, S.L.; Batra, J. Long Non-Coding RNAs at the Chromosomal Risk Loci Identified by Prostate and Breast Cancer GWAS. Genes 2021, 12, 2028. [Google Scholar] [CrossRef]
- Minotti, L.; Agnoletto, C.; Baldassari, F.; Corrà, F.; Volinia, S. SNPs and Somatic Mutation on Long Non-Coding RNA: New Frontier in the Cancer Studies? High Throughput 2018, 7, 34. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef]
- Botti, G.; Collina, F.; Scognamiglio, G.; Aquino, G.; Cerrone, M.; Liguori, G.; Gigantino, V.; Malzone, M.G.; Cantile, M. LncRNA HOTAIR Polymorphisms Association with Cancer Susceptibility in Different Tumor Types. Curr. Drug Targets 2018, 19, 1220–1226. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Y.; Li, Y.; Lin, F.; Zhang, J. Association between HOTAIR genetic polymorphisms and cancer susceptibility: A meta-analysis involving 122,832 subjects. Genomics 2020, 112, 3036–3055. [Google Scholar] [CrossRef]
- Kim, J.O.; Jun, H.H.; Kim, E.J.; Lee, J.Y.; Park, H.S.; Ryu, C.S.; Kim, S.; Oh, D.; Kim, J.W.; Kim, N.K. Genetic Variants of HOTAIR Associated with Colorectal Cancer Susceptibility and Mortality. Front. Oncol. 2020, 10, 72. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Yu, Q.-M.; Du, Y.-A.; Yang, L.-T.; Dong, R.-Z.; Huang, L.; Yu, P.-F.; Cheng, X.-D. Knockdown of Long Non-coding RNA HOTAIR Suppresses Tumor Invasion and Reverses Epithelial-mesenchymal Transition in Gastric Cancer. Int. J. Biol. Sci. 2013, 9, 587–597. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed. Pharmacother. 2017, 90, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef] [PubMed]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-J.; Sun, M.; Li, S.-Q.; Wu, Q.-Q.; Ji, L.; Liu, Z.-L.; Zhou, G.-Z.; Cao, G.; Jin, L.; Xie, H.-W.; et al. Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol. Carcinog. 2013, 52, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Z.; Mei, Q.; Guo, M.; Fu, X.; Han, W. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 109, 2266–2278. [Google Scholar] [CrossRef]
- Lv, X.-B.; Lian, G.-Y.; Wang, H.-R.; Song, E.; Yao, H.; Wang, M.-H. Long Noncoding RNA HOTAIR Is a Prognostic Marker for Esophageal Squamous Cell Carcinoma Progression and Survival. PLoS ONE 2013, 8, e63516. [Google Scholar] [CrossRef]
- Svoboda, M.; Slyskova, J.; Schneiderova, M.; Makovicky, P.; Bielik, L.; Levy, M.; Lipska, L.; Hemmelova, B.; Kala, Z.; Protivankova, M.; et al. Hotair long non-coding rna is anegative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014, 35, 1510–1515. [Google Scholar] [CrossRef]
- Wu, Z.H.; Wang, X.L.; Tang, H.M.; Jiang, T.; Chen, J.; Lu, S.; Qiu, G.Q.; Peng, Z.H.; Yan, D.W. Long non-coding rna hotair isa powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 2014, 32, 395–402. [Google Scholar] [CrossRef]
- Xue, Y.; Gu, D.; Ma, G.; Zhu, L.; Hua, Q.; Chu, H.; Tong, N.; Chen, J.; Zhang, Z.; Wang, M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis 2015, 30, 303–310. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; You, L.H.; Zhou, R.Z. Significant association between longnon-coding RNA HOTAIR polymorphisms and cancer susceptibility: A meta-analysis. OncoTargets Ther. 2016, 9, 3335–3343. [Google Scholar] [CrossRef]
- Qiu, H.; Liu, Q.; Li, J.; Wang, X.; Wang, Y.; Yuan, Z.; Li, J.; Pei, D.-S. Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. Apmis 2016, 124, 567–573. [Google Scholar] [CrossRef]
- Rajagopal, T.; Seshachalam, A.; Akshaya, R.; Rathnam, K.K.; Talluri, S.; Jothi, A.; Dunna, N.R. Association of HOTAIR (rs920778 and rs1899663) and NME1 (rs16949649 and rs2302254) gene polymorphisms with breast cancer risk in India. Gene 2020, 762, 145033. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Li, Y.-W.; Zhang, H.-B.; Gong, H.; Yuan, Y.; Li, W.-T.; Liu, H.-Y.; Chen, J. HOTAIR lncRNA SNPs rs920778 and rs1899663 are associated with smoking, male gender, and squamous cell carcinoma in a Chinese lung cancer population. Acta Pharmacol. Sin. 2018, 39, 1797–1803. [Google Scholar] [CrossRef]
- Lv, Z.; Kou, C.; Chen, N.; Jia, L.; Sun, X.; Gao, Y.; Bai, R.; Yang, M.; Cui, J. Single Nucleotide Polymorphisms in HOTAIR Are Related to Breast Cancer Risk and Prognosis in the Northeastern Chinese Population. Front. Oncol. 2021, 11, 706428. [Google Scholar] [CrossRef]
- Kashani, S.; Sasan, H.A.; Bahari, G.; Mollashahi, B.; Hashemi, S.M.; Taheri, M. Association between HOTAIR Polymorphisms and Lymphoma. Asian Pac. J. Cancer Prev. 2021, 22, 2831–2835. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Majello, B.; Gorini, F.; Saccà, C.D.; Amente, S. Expanding the Role of the Histone Lysine-Specific Demethylase LSD1 in Cancer. Cancers 2019, 11, 324. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Ishibashi, M.; Kogo, R.; Shibata, K.; Sawada, G.; Takahashi, Y.; Kurashige, J.; Akiyoshi, S.; Sasaki, S.; Iwaya, T.; Sudo, T.; et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol. Rep. 2013, 29, 946–950. [Google Scholar] [CrossRef]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.M.; McIver, C.M.; Stephenson, S.-A.; Hewett, P.J.; Rieger, N.; Hardingham, J.E. Identification of Early-Stage Colorectal Cancer Patients at Risk of Relapse Post-Resection by Immunobead Reverse Transcription-PCR Analysis of Peritoneal Lavage Fluid for Malignant Cells. Clin. Cancer Res. 2006, 12, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Lien, G.-S.; Lin, C.-H.; Yang, Y.-L.; Wu, M.-S.; Chen, B.-C. Ghrelin induces colon cancer cell proliferation through the GHS-R, Ras, PI3K, Akt, and mTOR signaling pathways. Eur. J. Pharmacol. 2016, 776, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Bayram, S.; Sümbül, A.T.; Batmacı, C.Y.; Genc, A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumor Biol. 2015, 36, 3863–3870. [Google Scholar] [CrossRef]
- Hassanzarei, S.; Hashemi, M.; Sattarifard, H.; Hashemi, S.M.; Bahari, G.; Ghavami, S. Genetic polymorphisms of HOTAIR gene are associated with the risk of breast cancer in a sample of southeast Iranian population. Tumor Biol. 2017, 39, 1010428317727539. [Google Scholar] [CrossRef]
- Taheri, M.; Habibi, M.; Noroozi, R.; Rakhshan, A.; Sarrafzadeh, S.; Sayad, A.; Omrani, M.D.; Ghafouri-Fard, S. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene 2017, 613, 20–24. [Google Scholar] [CrossRef]
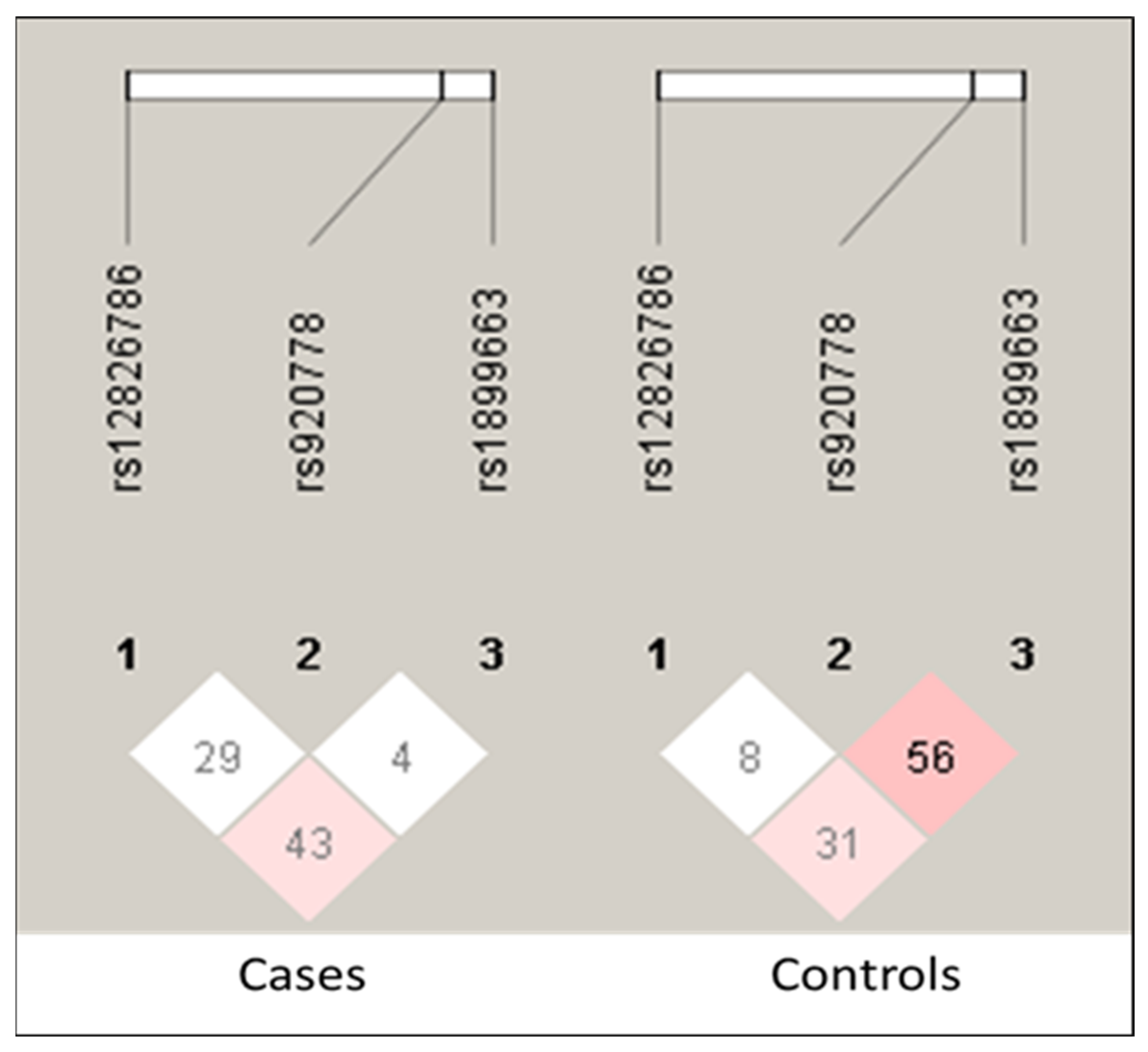
| Characteristics | Cases, n (%) | Control, n (%) |
|---|---|---|
| (n = 144) | (n = 144) | |
| Age in years (mean ± SD) | ||
| ≤57 | 68 (47%) | 78 (54.17%) |
| >57 | 76 (52.7%) | 66 (45.83%) |
| Sex | ||
| Males | 86 (59.7%) | 82 (56.94%) |
| Females | 58 (40%) | 62 (43.06%) |
| Tumor location | ||
| Colon | 91 (63%) | |
| Rectum | 53 (36.8%) | |
| Tumor node metastasis | ||
| Stage I-II | 61 (54%) | |
| Stage III-IV | 51 (46%) |
| Genotype | Controls | Cases | OR (95% CI) | χ2 | p |
|---|---|---|---|---|---|
| (n = 144) | (n = 144) | ||||
| rs920778 G > A | |||||
| GG | 48 (33%) | 35 (24%) | 1.000 (reference) | ||
| GA | 70 (49%) | 70 (49%) | 1.371 (0.793–2.371) | 1.28 | 0.25741 |
| AA | 26 (18%) | 39 (27%) | 2.057 (1.063–3.981) | 4.64 | 0.03131 |
| GA + AA | 96 (67%) | 109 (76%) | 1.557 (0.931–2.606) | 2.86 | 0.09078 |
| G | 83 (58%) | 70 (49%) | |||
| A | 61 (42%) | 74 (51%) | 1.438 (1.035–1.998) | 4.71 | 0.02994 |
| rs1899663 C > A | |||||
| CC | 10 (7%) | 16 (11%) | 1.000 (reference) | ||
| CA | 64 (44%) | 7 (5%) | 0.068 (0.023–0.208) | 28.1 | 1.153 |
| AA | 69 (48%) | 58 (40%) | 0.525 (0.221–1.246) | 2.18 | 0.14015 |
| CA + AA | 133 (92%) | 65 (45%) | 0.305 (0.131–0.710 | 8.21 | 0.00417 |
| C | 42 (29%) | 19.5 (14%) | |||
| A | 101 (70%) | 61.5 (43%) | 1.312 (0.844–2.038) | 1.46 | 0.22746 |
| rs12826786C > T | |||||
| CC | 93 (65%) | 92 (64%) | 1.000 (reference) | ||
| CT | 40 (28%) | 49 (34%) | 1.238 (0.746–2.057) | 0.68 | 0.40872 |
| TT | 11 (8%) | 3 (2%) | 0.276 (0.074–1.020) | 4.18 | 0.04094 |
| CT + TT | 51 (35%) | 52 (36%) | 1.031 (0.637–1.669) | 0.02 | 0.90215 |
| C | 133 (92%) | 116.5 (81%) | |||
| T | 31 (22%) | 27.5 (19%) | 0.86 (0.573–1.292) | 0.53 | 0.46848 |
| Genotype | ≤57 | >57 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | OR (95% CI) | χ2 | p | Controls | Cases | OR (95% CI) | χ2 | p | |
| (n = 78) | (n = 68) | (n = 66) | (n = 76) | |||||||
| rs920778 G > A | ||||||||||
| GG | 26 (33%) | 15 (22%) | 1.000 (reference) | 22 (33%) | 20 (26%) | 1.000 (reference) | ||||
| GA | 31 (40%) | 29 (43%) | 1.622 (0.720–3.654) | 1.4 | 0.24 | 39 (59%) | 41 (54%) | 1.156 (0.548–2.442) | 0.15 | 0.703 |
| AA | 21 (27%) | 24 (35%) | 1.981 (0.835–4.701) | 2.4 | 0.12 | 5 (8%) | 15 (20%) | 3.3 (1.015–10.733) | 4.13 | 0.043 |
| GA + AA | 52 (67%) | 53 (78%) | 1.767 (0.841–3.709) | 2.3 | 0.13 | 44 (67%) | 56 (74%) | 1.4 (0.679–2.885) | 0.84 | 0.361 |
| G | 28.5 (37%) | 29.5 (43%) | 30.5 (46%) | 40.5 (53%) | ||||||
| A | 36.5 (47%) | 38.5 (57%) | 1.484 (0.934–2.356) | 2.8 | 0.09 | 24.5 (37%) | 35.5 (47%) | 1.485 (0.923–2.389) | 2.66 | 0.103 |
| rs1899663 C > A | ||||||||||
| CC | 4 (5%) | 11 (16%) | 1.000 (reference) | 6 (9%) | 5 (7%) | 1.000 (reference) | ||||
| CA | 41 (53%) | 26 (38%) | 0.231 (0.066–0.801) | 5.9 | 0.0151 | 23 (35%) | 44 (58%) | 2.296 (0.632–8.336) | 1.65 | 0.198 |
| AA | 33 (42%) | 31 (46%) | 0.342 (0.098–1.186) | 3 | 0.08 | 36 (55%) | 27 (36%) | 0.9 (0.248–3.261) | 0.03 | 0.873 |
| CA + AA | 74 (95%) | 57 (84%) | 0.28 (0.085–0.926) | 4.81 | 0.0289 | 59 (89%) | 71 (93%) | 1.444 (0.420–4.970) | 0.34 | 0.558 |
| C | 24.5 (31%) | 24 (35%) | 17.5 (27%) | 27 (36%) | ||||||
| A | 53.5 (69%) | 44 (65%) | 0.84 (0.515–1.367) | 0.5 | 0.48 | 47.5 (72%) | 49 (64%) | 0.669 (0.401–1.114) | 2.4 | 0.121 |
| rs12826786C > T | ||||||||||
| CC | 54 (69%) | 45 (66%) | 1.000 (reference) | 39 (59%) | 47 (62%) | 1.000 (reference) | ||||
| CT | 18 (23%) | 21 (31%) | 1.4 (0.666–2.945) | 0.8 | 0.37 | 22 (33%) | 28 (37%) | 1.056 (0.524–2.130) | 0.02 | 0.879 |
| TT | 6 (8%) | 2 (3%) | 0.4 (0.077–2.080) | 1.3 | 0.26 | 5 (8%) | 1 (1%) | 0.166 (0.019–1.481) | 3.24 | 0.072 |
| CT + TT | 24 (31%) | 23 (34%) | 1.15 (0.574–2.305) | 0.2 | 0.69 | 27 (41%) | 29 (38%) | 0.891 (0.454–1.750) | 0.11 | 0.738 |
| C | 63 (81%) | 55.5 (82%) | 50 (76%) | 61 (80%) | ||||||
| T | 15 (19%) | 12.5 (18%) | 0.946 (0.525–1.705) | 0 | 0.85 | 16 (24%) | 15 (20%) | 0.768 (0.437–1.351) | 0.84 | 0.359 |
| Genotype | Males | Females | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | OR (95% CI) | χ2 | p | Controls | Cases | OR (95% CI) | χ2 | p | |
| (n = 82) | (n = 86) | (n = 62) | (n = 58) | |||||||
| rs920778 G > A | ||||||||||
| GG | 29 (35%) | 24 (28%) | 1.000 (reference) | 19 (31%) | 11 (19%) | 1.000 (reference) | ||||
| GA | 37 (45%) | 42 (49%) | 1.37 (0.682–2.758) | 0.79 | 0.375 | 33 (53%) | 28 (48%) | 1.466 (0.598–3.595) | 0.7 | 0.403 |
| AA | 16 (20%) | 20 (23%) | 1.51 (0.645–3.538) | 0.91 | 0.341 | 10 (16%) | 19 (33%) | 3.282 (1.129–9.536) | 4.91 | 0.0266 |
| GA + AA | 53 (65%) | 62 (72%) | 1.41 (0.735–2.717) | 1.08 | 0.298 | 43 (69%) | 47 (81%) | 1.888 (0.807–4.417) | 2.18 | 0.14 |
| G | 47.5 (58%) | 45 (52%) | Ref | 35.5 (57%) | 15 (26%) | |||||
| A | 34.5 (42%) | 41 (48%) | 1.25 (0.815–1.930) | 1.06 | 0.302 | 26.5 (43%) | 33 (57%) | 1.768 (1.060–2.949) | 4.8 | 0.0284 |
| rs1899663 C > A | ||||||||||
| CC | 6 (7%) | 8 (9%) | 1.000 (reference) | 4 (6%) | 8 (14%) | 1.000 (reference) | ||||
| CA | 33 (40%) | 37 (43%) | 0.84 (0.264–2.677) | 0.09 | 0.769 | 31 (50%) | 33 (57%) | 0.532 (0.146–1.946) | 0.93 | 0.335 |
| AA | 42 (51%) | 41 (48%) | 0.73 (0.234–2.295) | 0.29 | 0.592 | 27 (44%) | 17 (29%) | 0.315 (0.082–1.208) | 3 | 0.083 |
| CA + AA | 75 (91%) | 78 (91%) | 0.78 (0.258–2.355) | 0.2 | 0.659 | 58 (94%) | 50 (86%) | 0.431 (0.122–1.517) | 1.79 | 0.18 |
| C | 22.5 (27%) | 26.5 (31%) | 19.5 (31%) | 24.5 (42%) | ||||||
| A | 58.5 (71%) | 59.5 (69%) | 0.86 (0.539–1.385) | 0.37 | 0.542 | 13.5 (22%) | 33.5 (58%) | 0.627 (0.370–1.064) | 3 | 0.083 |
| rs12826786C > T | ||||||||||
| CC | 56 (68%) | 61 (71%) | 1.000 (reference) | 37 (60%) | 31 (53%) | 1.000 (reference) | ||||
| CT | 18 (22%) | 23 (27%) | 1.17 (0.574–2.399) | 0.19 | 0.662 | 22 (35%) | 26 (45%) | 1.411 (0.672–2.961) | 0.83 | 0.363 |
| TT | 8 (10%) | 2 (2%) | 0.23 (0.047–1.127) | 3.81 | 0.05107 | 3 (5%) | 1 (2%) | 0.398 (0.039–4.020) | 0.65 | 0.421 |
| CT + TT | 26 (32%) | 25 (29%) | 0.88 (0.457–1.704) | 0.14 | 0.71 | 25 (40%) | 27 (47%) | 1.289 (0.625–2.658 | 0.47 | 0.491 |
| C | 65 (79%) | 72.5 (84%) | 48 (77%) | 44 (76%) | ||||||
| T | 17 (21%) | 13.5 (16%) | 0.71 (0.408–1.244) | 1.43 | 0.231 | 14 (23%) | 13 (22%) | 1.091 (0.600–1.985) | 0.08 | 0.776 |
| Genotype | Control | Colon | OR (95% CI) | χ2 | p | Rectum | OR (95% CI) | χ2 | p |
|---|---|---|---|---|---|---|---|---|---|
| (n = 144) | (n = 91) | (n = 53) | |||||||
| rs920778 G > A | |||||||||
| GG | 48 (33%) | 19 (21%) | 1.000 (reference) | 16 (30%) | 1.000 (reference) | ||||
| GA | 70 (49%) | 48 (53%) | 1.7 (0.908–3.305) | 2.81 | 0.0938 | 22 (42%) | 0.9 (0.449–1.979) | 0.02 | 0.403 |
| AA | 26 (18%) | 24 (26%) | 2.3 (1.082–5.027) | 4.75 | 0.02926 | 15 (28%) | 1.7 (0.739–4.053) | 1.61 | 0.0266 |
| GA + AA | 96 (67%) | 72 (79%) | 1.8 (1.027–3.497) | 4.24 | 0.04209 | 37 (70%) | 1.15 (0.585–2.285) | 0.17 | 0.14 |
| G | 83 (58%) | 43 (47%) | 27 (51%) | ||||||
| A | 61 (42%) | 48 (53%) | 1.51 (1.046–2.206) | 4.84 | 0.02785 | 26 (49%) | 1.3 (0.838–2.048) | 1.41 | 0.0284 |
| rs1899663 C > A | |||||||||
| CC | 10 (7%) | 16 (11%) | 1.000 (reference) | 4 (8%) | 1.000 (reference) | ||||
| CA | 64 (44%) | 7 (5%) | 0.58 (0.233–1.473) | 1.31 | 0.2525 | 25 (47%) | 0.97 (0.280–3.403) | 0 | 0.97 |
| AA | 69 (48%) | 58 (40%) | 0.41 (0.161–1.045) | 3.61 | 0.05726 | 25 (47%) | 0.87 (0.260–3.151) | 0.02 | 0.88 |
| CA + AA | 133 (92%) | 65 (45%) | 0.49 (0.204–1.198) | 2.5 | 0.1135 | 50 (94%) | 0.91 (0.282–3.134) | 0.01 | 0.92 |
| C | 42 (29%) | 19.5 (14%) | 16.5 (31%) | ||||||
| A | 101 (70%) | 61.5 (43%) | 0.68 (0.460–1.009) | 3.69 | 0.05482 | 37.5 (71%) | 0.9 (0.584–1.530) | 0.05 | 0.82 |
| rs12826786C > T | |||||||||
| CC | 93 (65%) | 92 (64%) | 1.000 (reference) | 35 (66%) | 1.000 (reference) | ||||
| CT | 40 (28%) | 49 (34%) | 1.3 (0.738–2.308) | 0.84 | 0.3591 | 17 (32%) | 1.129 (0.568–2.247) | 0.12 | 0.73 |
| TT | 11 (8%) | 3 (2%) | 0.29 (0.063–1.387) | 2.65 | 0.1036 | 1 (2%) | 0.242 (0.030–1.941) | 2.08 | 0.15 |
| CT + TT | 51 (35%) | 52 (36%) | 1.08 (0.631–1.876) | 0.09 | 0.7623 | 18 (34%) | 0.93 (0.483–1.820) | 0.04 | 0.85 |
| C | 133 (92%) | 116.5 (81%) | 43.5 (82%) | ||||||
| T | 31 (22%) | 27.5 (19%) | 0.89 (0.567–1.424) | 0.21 | 0.6496 | 9.5 (18%) | 0.79 (0.450–1.408) | 0.62 | 0.43 |
| Genotype | Controls | Stage I-II (n = 61) | OR (95% CI) | χ2 | p | Stage I-II | OR (95% CI) | χ2 | p |
|---|---|---|---|---|---|---|---|---|---|
| (n = 144) | (n = 51) | ||||||||
| rs920778 G > A | |||||||||
| GG | 48 (33%) | 12 (20%) | 1.000 (reference) | 13 (25%) | 1.000 (reference) | ||||
| GA | 70 (49%) | 32 (52%) | 1.829 (0.857–3.90) | 2.47 | 0.11606 | 22 (43%) | 1.16 (0.533–2.526) | 0.14 | 0.70757 |
| AA | 26 (18%) | 17 (28%) | 2.615 (1.085–6.304) | 4.73 | 0.02972 | 16 (31%) | 2.272 (0.948–5.44) | 3.46 | 0.06272 |
| GA + AA | 96 (67%) | 49 (80%) | 2.042 (0.994–4.195) | 3.86 | 0.04937 | 38 (75%) | 1.462 (0.712–2.999) | 1.08 | 0.2992 |
| G | 83 (58%) | 28 (46%) | 24 (47%) | ||||||
| A | 61 (42%) | 33 (54%) | 1.604 (1.047–2.455) | 4.76 | 0.02921 | 27 (53%) | 1.531 (0.972–2.499) | 3.4 | 0.065 |
| rs1899663 C > A | |||||||||
| CC | 10 (7%) | 4 (7%) | 1.000 (reference) | 8 (16%) | 1.000 (reference) | ||||
| CA | 64 (44%) | 26 (43%) | 1.016 (0.292–3.530) | 0 | 0.98054 | 24 (47%) | 0.469 (0.165–1.328) | 2.09 | 0.14819 |
| AA | 69 (48%) | 31 (51%) | 1.123 (0.327–3.860) | 0.03 | 0.85361 | 19 (37%) | 0.344 (0.119–0.993) | 4.11 | 0.0426 |
| CA + AA | 133 (92%) | 57 (93%) | 1.071 (0.323–3.558) | 0.01 | 0.91029 | 43 (84%) | 0.0404 (0.150–1.089) | 3.38 | 0.06619 |
| C | 42 (29%) | 17 (28%) | 20 (39%) | ||||||
| A | 101 (70%) | 44 (72%) | 1.076 (0.672–1.723) | 0.09 | 0.75937 | 31 (61%) | 0.645 (0.402–1.033) | 3.35 | 0.06715 |
| rs12826786C > T | |||||||||
| CC | 93 (65%) | 43 (70%) | 1.000 (reference) | 32 (63%) | 1.000 (reference) | ||||
| CT | 40 (28%) | 17 (28%) | 0.919 (0.469–1.801) | 0.06 | 0.80605 | 19 (37%) | 1.38 (0.701–2.719) | 0.87 | 0.3503 |
| TT | 11 (8%) | 1 (2%) | 0.197 (0.025–1.572) | 2.86 | 0.09071 | 0% | 0.125 (0.007–2.183) | 3.68 | 0.05499 |
| CT + TT | 51 (35%) | 18 (30%) | 0.763 (0.399–1.459) | 0.67 | 0.41309 | 19 (37%) | 1.083 (0.558–2.100) | 0.06 | 0.81408 |
| C | 133 (92%) | 51.5 (84%) | 41.5 (81%) | ||||||
| T | 31 (22%) | 9.5 (16%) | 0.672 (0.382–1.182) | 1.92 | 0.16626 | 9.5 (19%) | 0.834 (0.471–1.479) | 0.39 | 0.53492 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alzeer, H.S.; Shaik, J.P.; Reddy Parine, N.; Alanazi, M.; Alamri, A.A.; Bhat, R.S.; Daihan, S.A. Genetic Variants of HOTAIR Associated with Colorectal Cancer: A Case-Control Study in the Saudi Population. Genes 2023, 14, 592. https://doi.org/10.3390/genes14030592
Alzeer HS, Shaik JP, Reddy Parine N, Alanazi M, Alamri AA, Bhat RS, Daihan SA. Genetic Variants of HOTAIR Associated with Colorectal Cancer: A Case-Control Study in the Saudi Population. Genes. 2023; 14(3):592. https://doi.org/10.3390/genes14030592
Chicago/Turabian StyleAlzeer, Haya Saad, Jilani P. Shaik, Narasimha Reddy Parine, Mohammad Alanazi, Abdullah Al Alamri, Ramesa Shafi Bhat, and Sooad Al Daihan. 2023. "Genetic Variants of HOTAIR Associated with Colorectal Cancer: A Case-Control Study in the Saudi Population" Genes 14, no. 3: 592. https://doi.org/10.3390/genes14030592
APA StyleAlzeer, H. S., Shaik, J. P., Reddy Parine, N., Alanazi, M., Alamri, A. A., Bhat, R. S., & Daihan, S. A. (2023). Genetic Variants of HOTAIR Associated with Colorectal Cancer: A Case-Control Study in the Saudi Population. Genes, 14(3), 592. https://doi.org/10.3390/genes14030592





