Novel Exon 7 Deletions in TSPAN12 in a Three-Generation FEVR Family: A Case Report and Literature Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Evaluation
2.2. Targeted Exome Sequencing of Genes for Inherited Eye Diseases and Sanger Sequencing
2.3. Whole-Genome Sequencing (WGS), CNV Analysis, and Fluorescent Quantitative PCR (qPCR)
2.4. Reverse Transcription PCR (RT-PCR) and Sequence Analysis
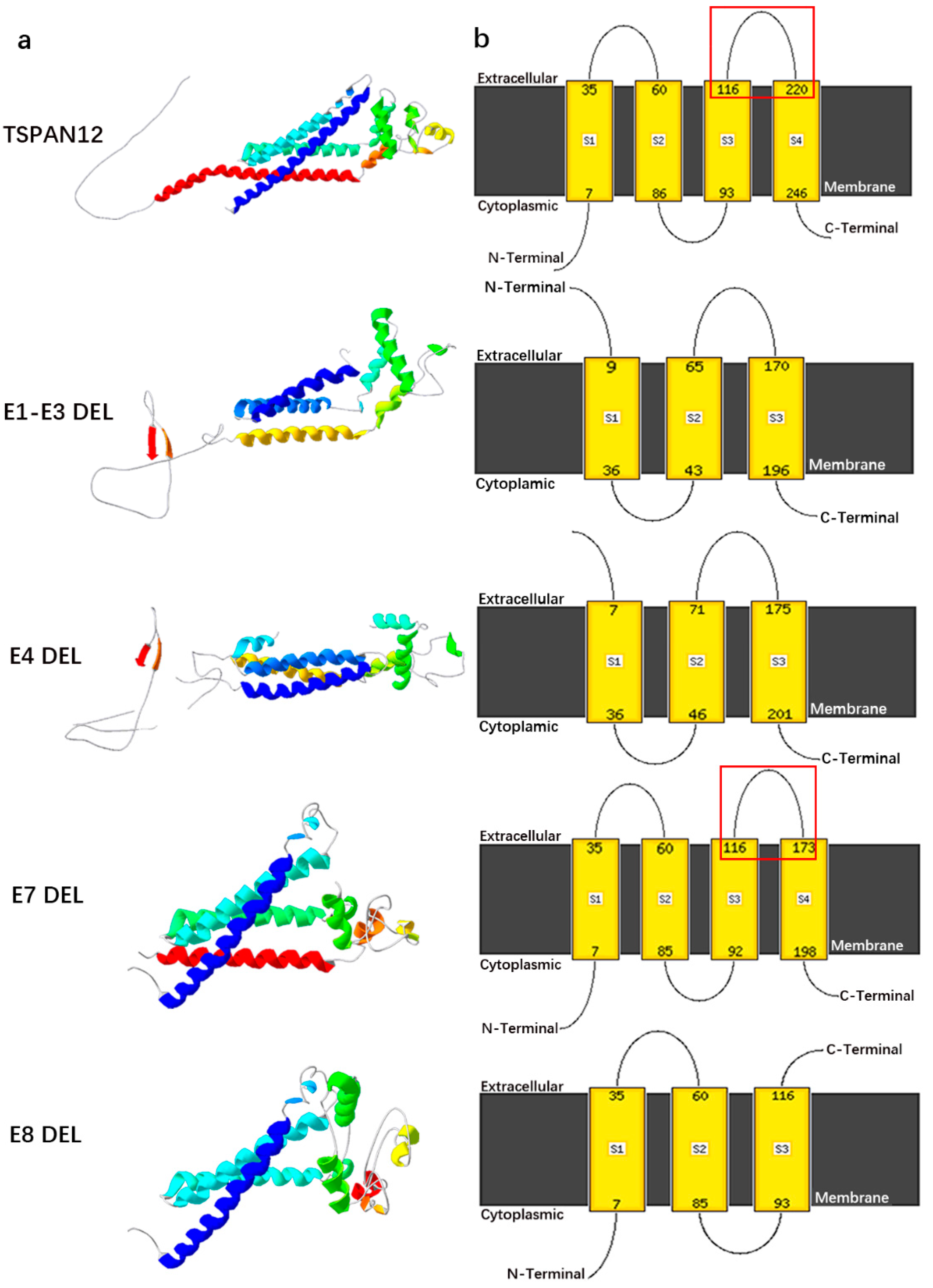
2.5. 3D Structure Modeling of TSPAN12 Protein after Gross Deletion
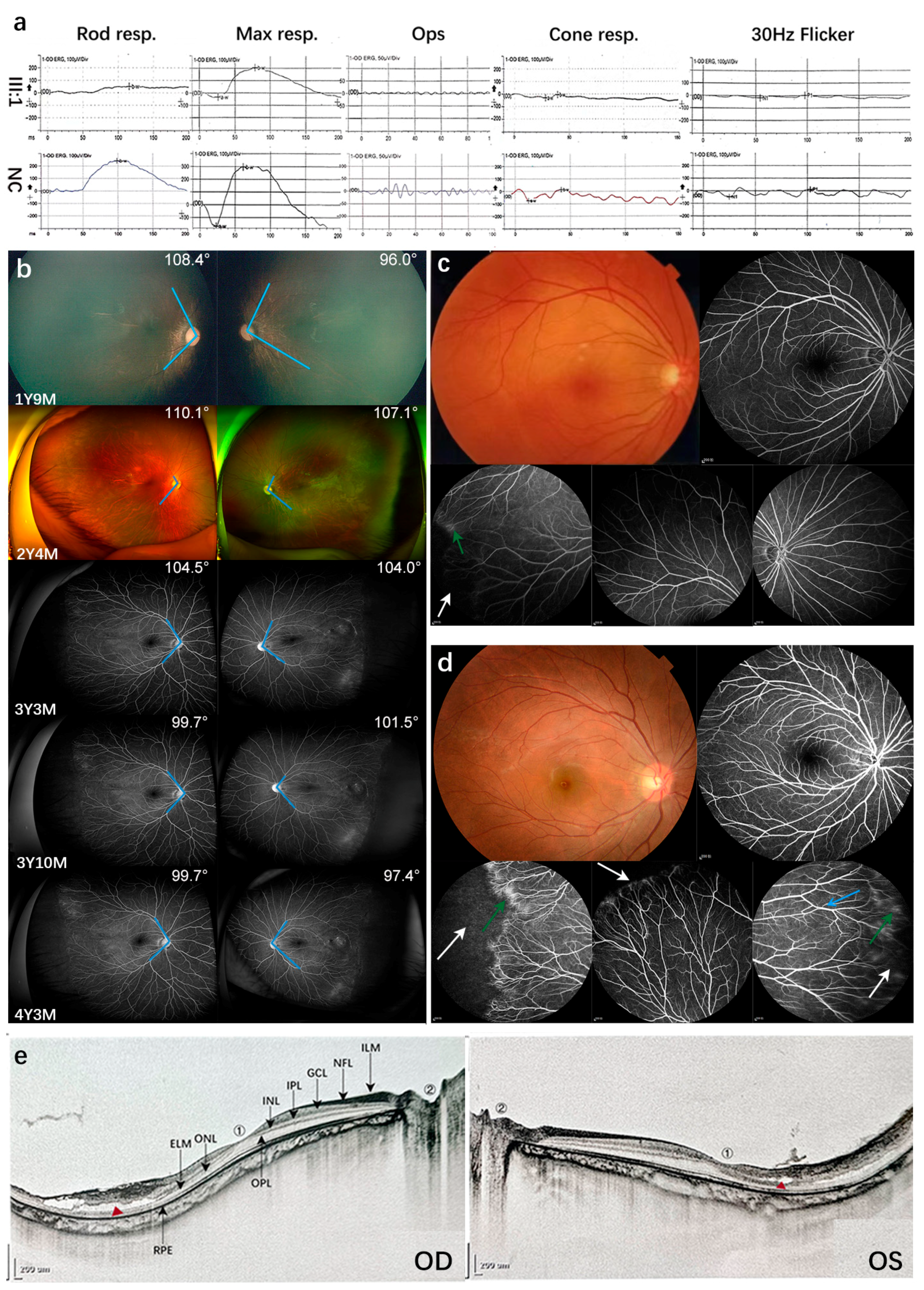
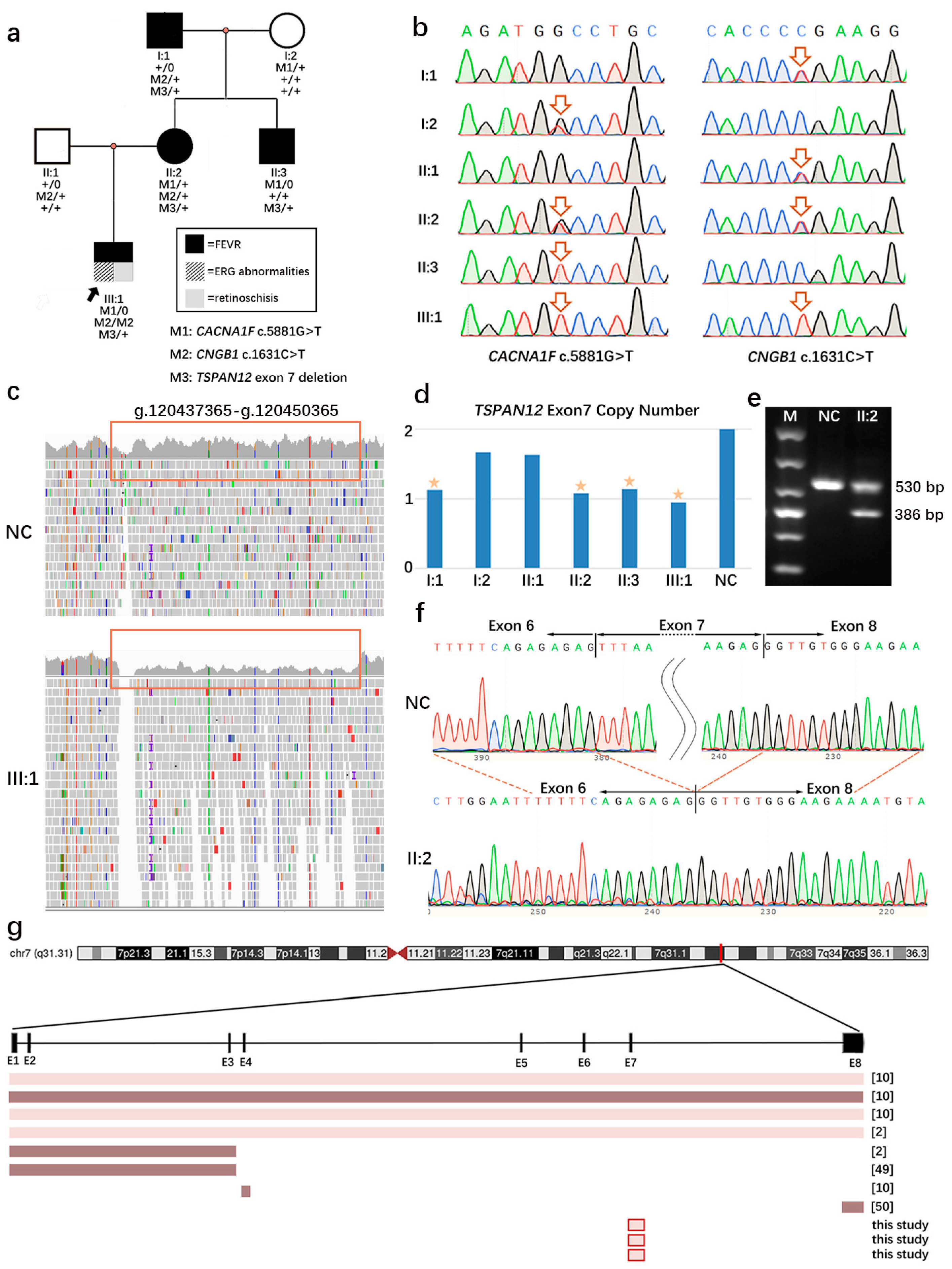
3. Results
3.1. Patient Characteristics and Genetic Analysis
3.2. Mild FEVR and Follow-Up Visits
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, H.; Li, N.; Li, Z.; Zhang, M.; Wei, M.; Huang, C.; Wang, J.; Li, F.; Wang, H.; Liu, Z.; et al. Fundus examination of 199 851 newborns by digital imaging in China: A multicentre cross-sectional study. Br. J. Ophthalmol. 2018, 102, 1742–1746. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Li, Y.; Zhang, X.; Chen, C.L.; Rao, Y.Q.; Fei, P.; Zhang, Q.; Zhao, P.; Li, J. Spectrum of Variants in 389 Chinese Probands With Familial Exudative Vitreoretinopathy. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5368–5381. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, M.; Huang, L.; Zhao, R.; Sundaresan, P.; Zhu, X.; Li, S.; Yang, Z. Whole-Exome Sequencing Reveals Novel TSPAN12 Variants in Autosomal Dominant Familial Exudative Vitreoretinopathy. Genet. Test. Mol. Biomark. 2021, 25, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rattner, A.; Zhou, Y.; Williams, J.; Smallwood, P.M.; Nathans, J. Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell 2012, 151, 1332–1344. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiao, X.; Li, S.; Mai, G.; Zhang, Q. Novel TSPAN12 mutations in patients with familial exudative vitreoretinopathy and their associated phenotypes. Mol. Vis. 2011, 17, 1128–1135. [Google Scholar] [PubMed]
- Ober, R.R.; Bird, A.C.; Hamilton, A.M.; Sehmi, K. Autosomal dominant exudative vitreoretinopathy. Br. J. Ophthalmol. 1980, 64, 112–120. [Google Scholar] [CrossRef] [PubMed]
- van Nouhuys, C.E. Signs, complications, and platelet aggregation in familial exudative vitreoretinopathy. Am. J. Ophthalmol. 1991, 111, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, S.D.; Trese, M.T. Familial exudative vitreoretinopathy. Results of surgical management. Ophthalmology 1998, 105, 1015–1023. [Google Scholar] [CrossRef]
- Chen, C.; Sun, L.; Li, S.; Huang, L.; Zhang, T.; Wang, Z.; Yu, B.; Ding, X. The spectrum of genetic mutations in patients with asymptomatic mild familial exudative vitreoretinopathy. Exp. Eye Res. 2020, 192, 107941. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Kim, M.J.; Park, S.W.; Kim, J.H.; Yu, Y.S.; Song, J.Y.; Cho, S.I.; Ahn, J.H.; Oh, Y.H.; Lee, J.S.; et al. Large Deletions of TSPAN12 Cause Familial Exudative Vitreoretinopathy (FEVR). Investig. Ophthalmol. Vis. Sci. 2016, 57, 6902–6908. [Google Scholar] [CrossRef]
- Tang, M.; Sun, L.; Hu, A.; Yuan, M.; Yang, Y.; Peng, X.; Ding, X. Mutation Spectrum of the LRP5, NDP, and TSPAN12 Genes in Chinese Patients With Familial Exudative Vitreoretinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5949–5957. [Google Scholar] [CrossRef]
- McCulloch, D.L.; Marmor, M.F.; Brigell, M.G.; Hamilton, R.; Holder, G.E.; Tzekov, R.; Bach, M. ISCEV Standard for full-field clinical electroretinography (2015 update). Doc. Ophthalmologica. Adv. Ophthalmol. 2015, 130, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, S.; Sun, W.; Xiao, X.; Jia, X.; Liu, M.; Xu, L.; Long, Y.; Zhang, Q. An Ophthalmic Targeted Exome Sequencing Panel as a Powerful Tool to Identify Causative Mutations in Patients Suspected of Hereditary Eye Diseases. Transl. Vis. Sci. Technol. 2019, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, D.; Xiao, X.; Li, S.; Jia, X.; Sun, W.; Guo, X.; Zhang, Q. Evaluation of 12 myopia-associated genes in Chinese patients with high myopia. Investig. Ophthalmol. Vis. Sci. 2015, 56, 722–729. [Google Scholar] [CrossRef]
- Li, R.; Shen, X.; Chen, H.; Peng, D.; Wu, R.; Sun, H. Developmental validation of the MGIEasy Signature Identification Library Prep Kit, an all-in-one multiplex system for forensic applications. Int. J. Legal Med. 2021, 135, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stutz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Geoffroy, V.; Herenger, Y.; Kress, A.; Stoetzel, C.; Piton, A.; Dollfus, H.; Muller, J. AnnotSV: An integrated tool for structural variations annotation. Bioinformatics 2018, 34, 3572–3574. [Google Scholar] [CrossRef] [PubMed]
- Rose-Zerilli, M.J.; Barton, S.J.; Henderson, A.J.; Shaheen, S.O.; Holloway, J.W. Copy-number variation genotyping of GSTT1 and GSTM1 gene deletions by real-time PCR. Clin. Chem. 2009, 55, 1680–1685. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Lindahl, E.; Wallner, B. Improved model quality assessment using ProQ2. BMC Bioinform. 2012, 13, 224. [Google Scholar] [CrossRef]
- Jones, D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 2007, 23, 538–544. [Google Scholar] [CrossRef]
- Kader, M.A. Electrophysiological study of myopia. Saudi J. Ophthalmol. 2012, 26, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Marr, J.E.; Halliwell-Ewen, J.; Fisher, B.; Soler, L.; Ainsworth, J.R. Associations of high myopia in childhood. Eye (Lond) 2001, 15, 70–74. [Google Scholar] [CrossRef]
- Xiao, X.; Jia, X.; Guo, X.; Li, S.; Yang, Z.; Zhang, Q. CSNB1 in Chinese families associated with novel mutations in NYX. J. Hum. Genet. 2006, 51, 634–640. [Google Scholar] [CrossRef]
- De Silva, S.R.; Arno, G.; Robson, A.G.; Fakin, A.; Pontikos, N.; Mohamed, M.D.; Bird, A.C.; Moore, A.T.; Michaelides, M.; Webster, A.R.; et al. The X-linked retinopathies: Physiological insights, pathogenic mechanisms, phenotypic features and novel therapies. Prog. Retin. Eye Res. 2021, 82, 100898. [Google Scholar] [CrossRef]
- Xu, X.; Li, S.; Xiao, X.; Wang, P.; Guo, X.; Zhang, Q. Sequence variations of GRM6 in patients with high myopia. Mol. Vis. 2009, 15, 2094–2100. [Google Scholar] [PubMed]
- Zeitz, C.; Jacobson, S.G.; Hamel, C.P.; Bujakowska, K.; Neuillé, M.; Orhan, E.; Zanlonghi, X.; Lancelot, M.E.; Michiels, C.; Schwartz, S.B.; et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2013, 92, 67–75. [Google Scholar] [CrossRef]
- Ahmad, N.N.; McDonald-McGinn, D.M.; Zackai, E.H.; Knowlton, R.G.; LaRossa, D.; DiMascio, J.; Prockop, D.J. A second mutation in the type II procollagen gene (COL2AI) causing stickler syndrome (arthro-ophthalmopathy) is also a premature termination codon. Am. J. Hum. Genet. 1993, 52, 39–45. [Google Scholar] [PubMed]
- Annunen, S.; Korkko, J.; Czarny, M.; Warman, M.L.; Brunner, H.G.; Kaariainen, H.; Mulliken, J.B.; Tranebjaerg, L.; Brooks, D.G.; Cox, G.F.; et al. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am. J. Hum. Genet. 1999, 65, 974–983. [Google Scholar] [CrossRef]
- Van Camp, G.; Snoeckx, R.L.; Hilgert, N.; van den Ende, J.; Fukuoka, H.; Wagatsuma, M.; Suzuki, H.; Smets, R.M.; Vanhoenacker, F.; Declau, F.; et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am. J. Hum. Genet. 2006, 79, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Booth, C.; Fillman, C.; Shapiro, M.; Blair, M.P.; Hyland, J.C.; Ala-Kokko, L. A loss of function mutation in the COL9A2 gene causes autosomal recessive Stickler syndrome. Am. J. Med. Genetics. Part A 2011, 155A, 1668–1672. [Google Scholar] [CrossRef]
- Dietz, H.C.; McIntosh, I.; Sakai, L.Y.; Corson, G.M.; Chalberg, S.C.; Pyeritz, R.E.; Francomano, C.A. Four novel FBN1 mutations: Significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics 1993, 17, 468–475. [Google Scholar] [CrossRef]
- Sun, W.; Huang, L.; Xu, Y.; Xiao, X.; Li, S.; Jia, X.; Gao, B.; Wang, P.; Guo, X.; Zhang, Q. Exome Sequencing on 298 Probands With Early-Onset High Myopia: Approximately One-Fourth Show Potential Pathogenic Mutations in RetNet Genes. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8365–8372. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Renner, A.B.; Kellner, U.; Fiebig, B.; Cropp, E.; Foerster, M.H.; Weber, B.H. ERG variability in X-linked congenital retinoschisis patients with mutations in the RS1 gene and the diagnostic importance of fundus autofluorescence and OCT. Doc. Ophthalmologica. Adv. Ophthalmol. 2008, 116, 97–109. [Google Scholar] [CrossRef]
- Arbabi, A.; Liu, A.; Ameri, H. Gene Therapy for Inherited Retinal Degeneration. J. Ocul. Pharmacol. Ther. 2019, 35, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Kusaka, S.; Ishimaru, M.; Kondo, H.; Kuniyoshi, K. Early vitrectomy to reverse macular dragging in a one-month-old boy with familial exudative vitreoretinopathy. Am. J. Ophthalmol. Case Rep. 2019, 15, 100493. [Google Scholar] [CrossRef] [PubMed]
- Tauqeer, Z.; Yonekawa, Y. Familial Exudative Vitreoretinopathy: Pathophysiology, Diagnosis, and Management. Asia Pac. J. Ophthalmol. (Phila) 2018, 7, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Reviews. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Tong, Y.; Zhu, Y.; Peng, M. Familial Exudative Vitreoretinopathy-Related Disease-Causing Genes and Norrin/beta-Catenin Signal Pathway: Structure, Function, and Mutation Spectrums. J. Ophthalmol. 2019, 2019, 5782536. [Google Scholar] [CrossRef] [PubMed]
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205. [Google Scholar] [CrossRef]
- Lai, M.B.; Zhang, C.; Shi, J.; Johnson, V.; Khandan, L.; McVey, J.; Klymkowsky, M.W.; Chen, Z.; Junge, H.J. TSPAN12 Is a Norrin Co-receptor that Amplifies Frizzled4 Ligand Selectivity and Signaling. Cell Rep. 2017, 19, 2809–2822. [Google Scholar] [CrossRef]
- Musada, G.R.; Syed, H.; Jalali, S.; Chakrabarti, S.; Kaur, I. Mutation spectrum of the FZD-4, TSPAN12 AND ZNF408 genes in Indian FEVR patients. BMC Ophthalmol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Luo, J.; Li, J.; Zhang, X.; Li, J.K.; Chen, H.J.; Zhao, P.Q.; Fei, P. Five novel copy number variations detected in patients with familial exudative vitreoretinopathy. Mol. vision 2021, 27, 632–642. [Google Scholar]
- Huang, X.Y.; Zhuang, H.; Wu, J.H.; Li, J.K.; Hu, F.Y.; Zheng, Y.; Tellier, L.; Zhang, S.H.; Gao, F.J.; Zhang, J.G.; et al. Targeted next-generation sequencing analysis identifies novel mutations in families with severe familial exudative vitreoretinopathy. Mol. Vis. 2017, 23, 605–613. [Google Scholar]
- Kashani, A.H.; Brown, K.T.; Chang, E.; Drenser, K.A.; Capone, A.; Trese, M.T. Diversity of retinal vascular anomalies in patients with familial exudative vitreoretinopathy. Ophthalmology 2014, 121, 2220–2227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Blair, M.P.; Zeid, J.L.; Basith, S.S.T.; Shapiro, M.J. FEVR phenotype associated with septo-optic dysplasia. Ophthalmic Genet. 2019, 40, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xiao, X.; Li, S.; Jia, X.; Wang, P.; Zhang, Q. Pathogenic variants and associated phenotypic spectrum of TSPAN12 based on data from a large cohort. Graefe′s Arch. Clin. Exp. 2021, 259, 2929–2939. [Google Scholar] [CrossRef] [PubMed]
- Kashani, A.H.; Learned, D.; Nudleman, E.; Drenser, K.A.; Capone, A.; Trese, M.T. High prevalence of peripheral retinal vascular anomalies in family members of patients with familial exudative vitreoretinopathy. Ophthalmology 2014, 121, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, L.; Li, J.; Zhang, Q.; Fei, P.; Zhu, X.; Tai, Z.; Ma, S.; Gong, B.; Li, Y.; et al. Novel mutations in the TSPAN12 gene in Chinese patients with familial exudative vitreoretinopathy. Mol. Vis. 2014, 20, 1296–1306. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Wang, P. Novel Exon 7 Deletions in TSPAN12 in a Three-Generation FEVR Family: A Case Report and Literature Review. Genes 2023, 14, 587. https://doi.org/10.3390/genes14030587
Jiang Z, Wang P. Novel Exon 7 Deletions in TSPAN12 in a Three-Generation FEVR Family: A Case Report and Literature Review. Genes. 2023; 14(3):587. https://doi.org/10.3390/genes14030587
Chicago/Turabian StyleJiang, Zixuan, and Panfeng Wang. 2023. "Novel Exon 7 Deletions in TSPAN12 in a Three-Generation FEVR Family: A Case Report and Literature Review" Genes 14, no. 3: 587. https://doi.org/10.3390/genes14030587
APA StyleJiang, Z., & Wang, P. (2023). Novel Exon 7 Deletions in TSPAN12 in a Three-Generation FEVR Family: A Case Report and Literature Review. Genes, 14(3), 587. https://doi.org/10.3390/genes14030587




