Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq
Abstract
:1. Introduction
2. Aim
3. Materials and Methods
3.1. In Vitro Embryo Production
3.2. Embryo Biopsy and cDNA Synthesis
3.3. WT RNA Sequencing
3.4. Mapping Raw RNA-Seq Data
3.5. RNA-Seq Statistical Analysis
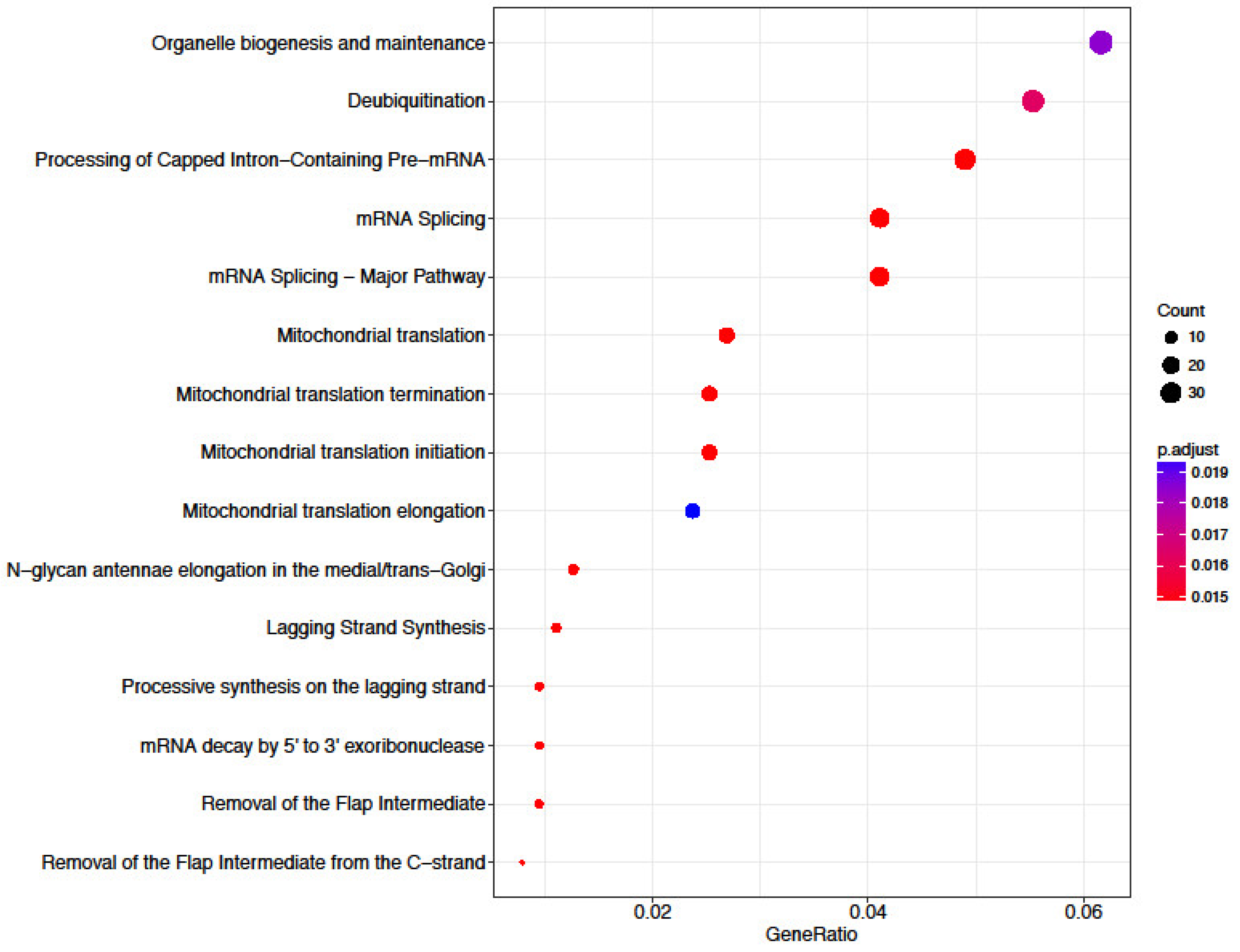
3.6. Functional Annotation of Differentially Expressed Genes
3.7. Quantitative Real-Time PCR Validation
3.8. Quantitative Real-Time PCR Statistical Analysis
4. Results
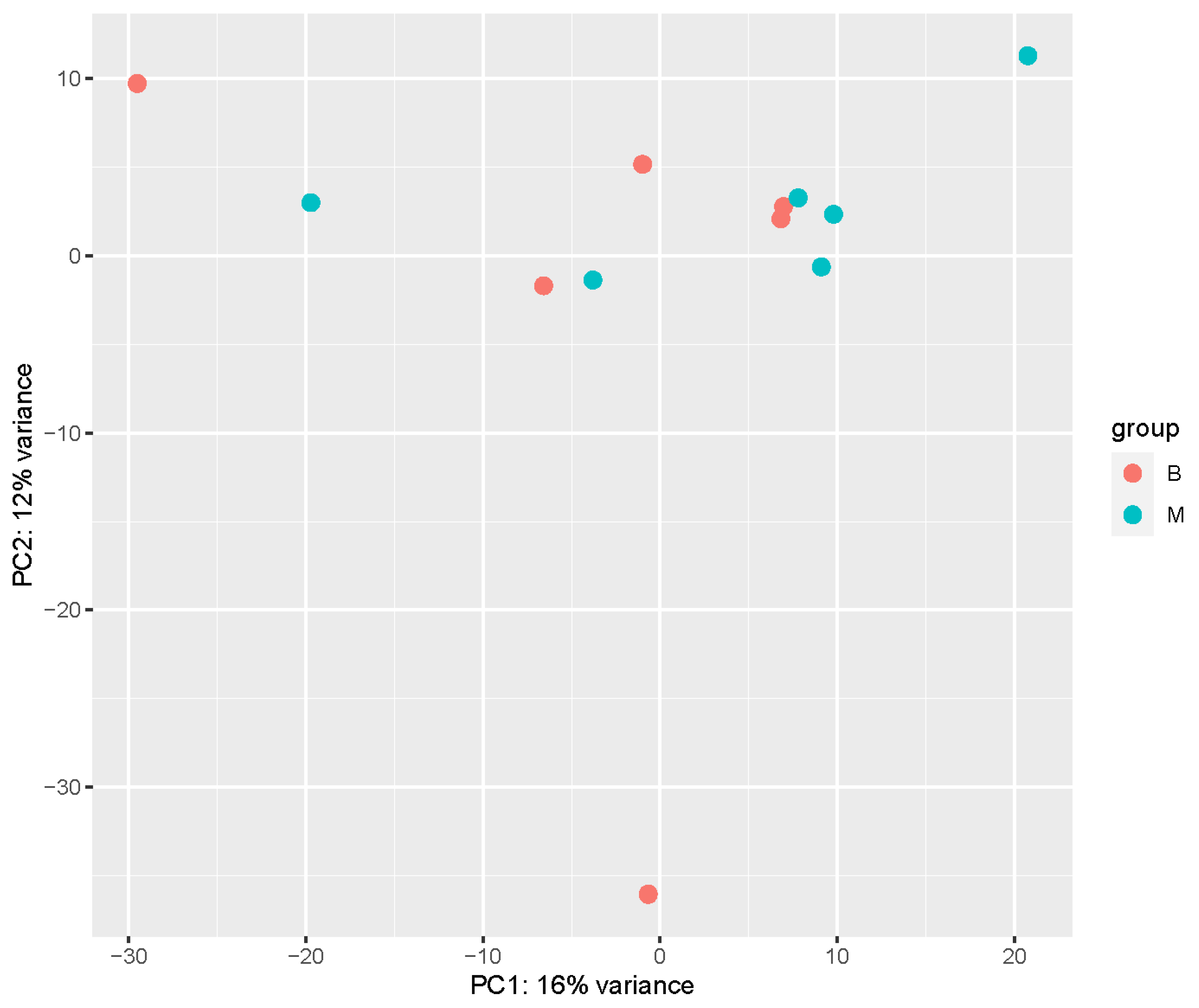
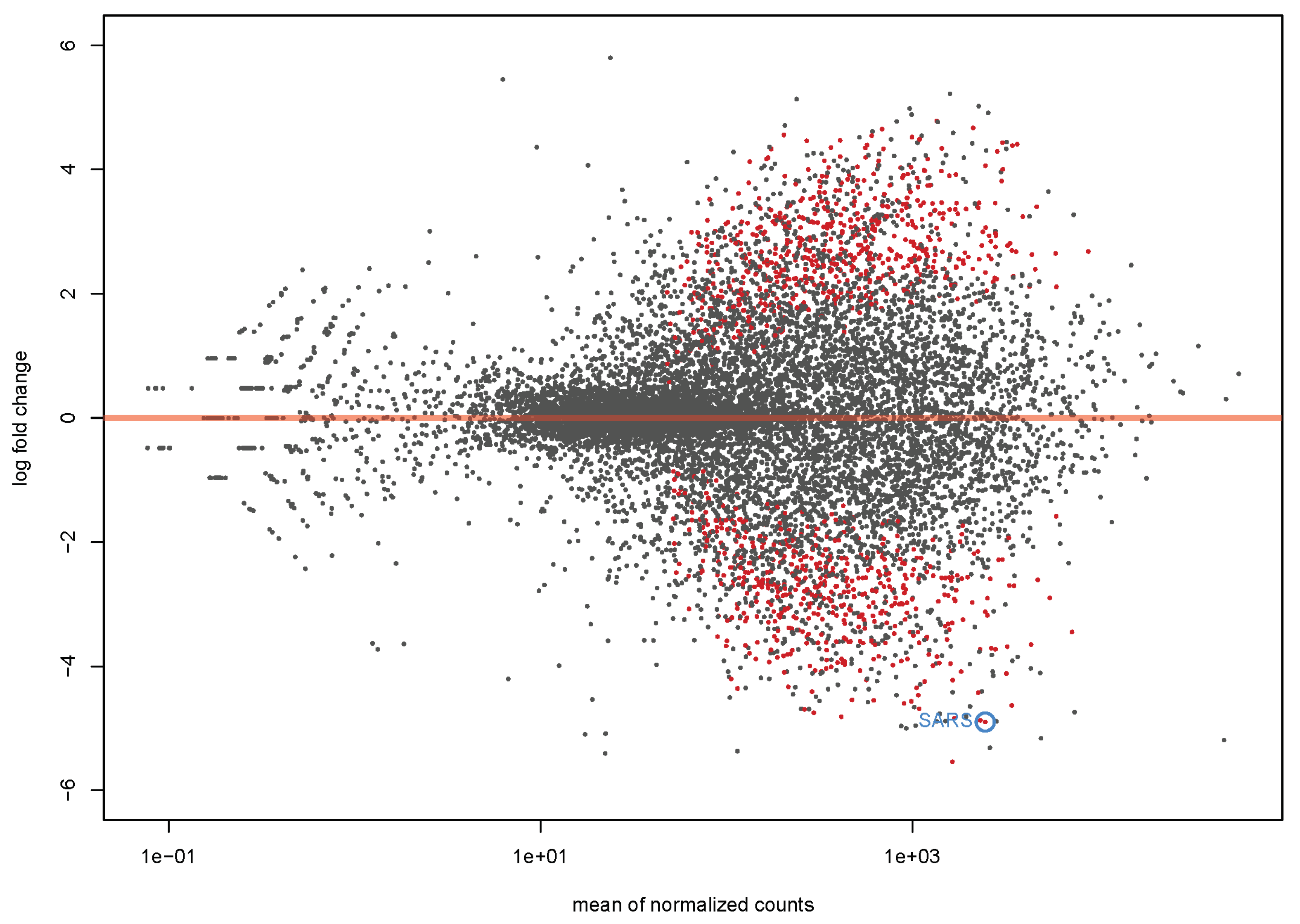
4.1. Transcriptome Analysis
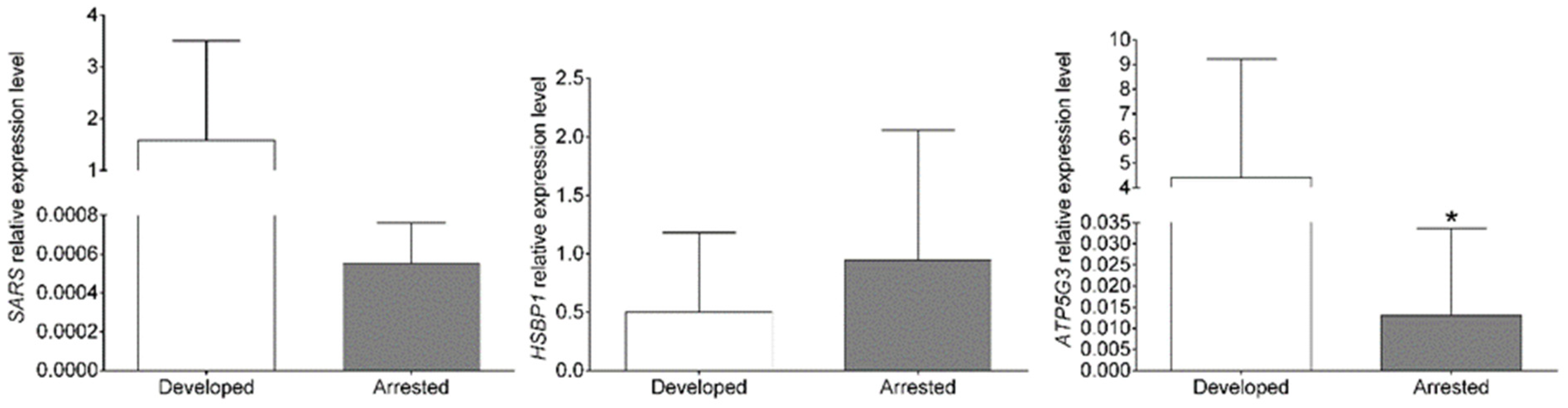
4.2. Quantitative Real-Time PCR Confirmation
5. Discussion
Study Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cenariu, M.; Pall, E.; Cernea, C.; Groza, I. Evaluation of Bovine Embryo Biopsy Techniques according to Their Ability to Preserve Embryo Viability. J. Biomed. Biotechnol. 2012, 2012, 541384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Sousa, R.V.; Cardoso, C.R.D.S.; Butzke, G.; Dode, M.A.N.; Rumpf, R.; Franco, M.M. Biopsy of bovine embryos produced in vivo and in vitro does not affect pregnancy rates. Theriogenology 2017, 90, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Forell, F.; Oliveira, A.; Rodrigues, J. Splitting and biopsy for bovine embryo sexing under field conditions. Theriogenology 2001, 56, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.S.; Quintão, C.C.R.; de Freitas, C.; Camargo, A.J.D.R.; Serapião, R.V.; Camargo, L.S.D.A. Post implantation development reveals that biopsy procedure can segregate “healthy” from “unhealthy” bovine embryos and prevent miscarriages. Anim. Reprod. Sci. 2017, 184, 51–58. [Google Scholar] [CrossRef]
- Polisseni, J.; de Sá, W.F.; Guerra, M.D.O.; Machado, M.A.; Serapião, R.V.; de Carvalho, B.C.; Camargo, L.S.D.A.; Peters, V.M. Post-biopsy bovine embryo viability and whole genome amplification in preimplantation genetic diagnosis. Fertil. Steril. 2010, 93, 783–788. [Google Scholar] [CrossRef]
- Bouquet, A.; Juga, J. Integrating genomic selection into dairy cattle breeding programmes: A review. Animal 2013, 7, 705–713. [Google Scholar] [CrossRef] [Green Version]
- Granleese, T.; Clark, S.A.; van der Werf, J.H.J. Genotyping strategies of selection candidates in livestock breeding programmes. J. Anim. Breed. Genet. 2019, 136, 91–101. [Google Scholar] [CrossRef]
- Jaton, C.; Schenkel, F.; Malchiodi, F.; Sargolzaei, M.; Price, C.; Baes, C.; Miglior, F. Genetic analysis for quality of frozen embryos produced by Holstein cattle donors in Canada. J. Dairy Sci. 2017, 100, 7320–7329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liang, D.; Huang, H.; Yang, Z.; Wang, Y.; Yu, Y.; Liu, L.; Zhang, S.; Han, J.; Xiao, W. Technical note: Development and application of KASP assays for rapid screening of 8 genetic defects in Holstein cattle. J. Dairy Sci. 2020, 103, 619–624. [Google Scholar] [CrossRef]
- Zacchini, F.; Arena, R.; Abramik, A.; Ptak, G.E. Embryo biopsy and development: The known and the unknown. Reproduction 2017, 154, R143–R148. [Google Scholar] [CrossRef] [Green Version]
- Cimadomo, D.; Capalbo, A.; Ubaldi, F.M.; Scarica, C.; Palagiano, A.; Canipari, R.; Rienzi, L. The Impact of Biopsy on Human Embryo Developmental Potential during Preimplantation Genetic Diagnosis. BioMed Res. Int. 2016, 2016, 7193075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimadomo, D.; Rienzi, L.; Capalbo, A.; Rubio, C.; Innocenti, F.; García-Pascual, C.M.; Ubaldi, F.M.; Handyside, A. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Hum. Reprod. Update 2020, 26, 453–473. [Google Scholar] [CrossRef] [PubMed]
- Rubio, C.; Rodrigo, L.; Garcia-Pascual, C.; Peinado, V.; Campos-Galindo, I.; Garcia-Herrero, S.; Simón, C. Clinical application of embryo aneuploidy testing by next-generation sequencing. Biol. Reprod. 2019, 101, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Robbins, K.M.; Wells, K.D.; Rivera, R.M. Large offspring syndrome. Epigenetics 2013, 8, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Ménézo, Y.J.; Hérubel, F. Mouse and bovine models for human IVF. Reprod. Biomed. Online 2002, 4, 170–175. [Google Scholar] [CrossRef]
- Sirard, M.-A. The influence of in vitro fertilization and embryo culture on the embryo epigenetic constituents and the possible consequences in the bovine model. J. Dev. Orig. Health Dis. 2017, 8, 411–417. [Google Scholar] [CrossRef]
- Avery, B.; Melsted, J.; Greve, T. A novel approach for in vitro production of bovine embryos: Use of the oxoid atmosphere generating system. Theriogenology 2000, 54, 1259–1268. [Google Scholar] [CrossRef]
- Bó, G.A.; Mapletoft, R.J. Evaluation and classification of bovine embryos. Anim. Reprod. 2013, 10, 344–348. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2015, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Chrenek, P.; Boulanger, L.; Heyman, Y.; Uhrin, P.; Laurincik, J.; Bulla, J.; Renard, J.-P. Sexing and multiple genotype analysis from a single cell of bovine embryo. Theriogenology 2001, 55, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Hamada, Y. Efficient production of sex-identified and cryosurvived bovine in-vitro produced blastocysts. Theriogenology 2004, 61, 1181–1191. [Google Scholar] [CrossRef]
- Tutt, D.A.; Passaro, C.; Whitworth, D.; Holland, M.K. Laser assisted blastomere extrusion biopsy of in vitro produced cattle embryos—A potential high throughput, minimally invasive approach for sampling pre-morula and morula stage embryos. Anim. Reprod. Sci. 2020, 219, 106546. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, K.; Nõmm, M.; Lättekivi, F.; Ressaissi, Y.; Godakumara, K.; Lavrits, A.; Midekessa, G.; Viil, J.; Bæk, R.; Jørgensen, M.M.; et al. Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non-invasive embryo quality markers. Theriogenology 2020, 149, 104–116. [Google Scholar] [CrossRef]
- Nõmm, M.; Porosk, R.; Pärn, P.; Kilk, K.; Soomets, U.; Kõks, S.; Jaakma, Ü. In vitro culture and non-invasive metabolic profiling of single bovine embryos. Reprod. Fertil. Dev. 2019, 31, 306. [Google Scholar] [CrossRef]
- Kubisch, H.; Johnson, K. The Effects of Blastomere Biopsy and Oxygen Tension on Bovine Embryo Development, Rate of Apoptosis and Interferon-τ Secretion. Reprod. Domest. Anim. 2007, 42, 509–515. [Google Scholar] [CrossRef]
- Orvieto, R.; Feldman, B.; Wiesel, M.; Shani, H.; Aizer, A. Is Day-4 morula biopsy a feasible alternative for preimplantation genetic testing? PLoS ONE 2020, 15, e0238599. [Google Scholar] [CrossRef]
- Zakharova, E.E.; Zaletova, V.V.; Krivokharchenko, A.S. Biopsy of Human Morula-Stage Embryos: Outcome of 215 IVF/ICSI Cycles with PGS. PLoS ONE 2014, 9, e106433. [Google Scholar] [CrossRef]
- Lammers, J.; Reignier, A.; Loubersac, S.; Chtourou, S.; Lefebvre, T.; Barrière, P.; Fréour, T. Modification of late human embryo development after blastomere removal on day 3 for preimplantation genetic testing. Syst. Biol. Reprod. Med. 2020, 67, 121–126. [Google Scholar] [CrossRef]
- Driver, A.M.; Khatib, H. PHYSIOLOGY AND ENDOCRINOLOGY SYMPOSIUM: Heat shock proteins: Potentially powerful markers for preimplantation embryonic development and fertility in livestock species1,2. J. Anim. Sci. 2013, 91, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wan, A.; Zhou, Z.; Chen, D.; Liang, H.; Liu, C.; Yan, S.; Niu, Y.; Lin, Z.; Zhan, S.; et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut 2020, 70, 1698–1712. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.; Swann, K. Mitochondria and lipid metabolism in mammalian oocytes and early embryos. Int. J. Dev. Biol. 2019, 63, 93–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, A.J. Mitochondria in early development: Linking the microenvironment, metabolism and the epigenome. Reproduction 2019, 157, R159–R179. [Google Scholar] [CrossRef] [Green Version]
- Van Blerkom, J. Mitochondria in early mammalian development. Semin. Cell Dev. Biol. 2009, 20, 354–364. [Google Scholar] [CrossRef]
- Van Blerkom, J. Mitochondria as regulatory forces in oocytes, preimplantation embryos and stem cells. Reprod. Biomed. Online 2008, 16, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Viotti, M. Mitochondrial DNA as a readout of embryo cellularity. J. Assist. Reprod. Genet. 2019, 36, 1845–1846. [Google Scholar] [CrossRef]
- Haouzi, D.; Boumela, I.; Chebli, K.; Hamamah, S. Global, Survival, and Apoptotic Transcriptome during Mouse and Human Early Embryonic Development. BioMed Res. Int. 2018, 2018, 5895628. [Google Scholar] [CrossRef] [Green Version]
- Lorda-Diez, C.I.; Garcia-Riart, B.; Montero, J.A.; Rodriguez-León, J.; A Garcia-Porrero, J.; Hurle, J.M. Apoptosis during embryonic tissue remodeling is accompanied by cell senescence. Aging 2015, 7, 974–985. [Google Scholar] [CrossRef] [Green Version]
- Tseng, Y.-C.; Cabot, B.; Cabot, R.A. ARID1A, a component of SWI/SNF chromatin remodeling complexes, is required for porcine embryo development. Mol. Reprod. Dev. 2017, 84, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. Sirt1 and the Mitochondria. Mol. Cells 2016, 39, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Adamkova, K.; Yi, Y.-J.; Petr, J.; Zalmanova, T.; Hoskova, K.; Jelinkova, P.; Moravec, J.; Kralickova, M.; Sutovsky, M.; Sutovsky, P.; et al. SIRT1-dependent modulation of methylation and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic pronuclei improves porcine embryo development. J. Anim. Sci. Biotechnol. 2017, 8, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aksu, K.; Golal, E.; Aslan, M.A.; Ustunel, I.; Acar, N. The investigation of the role of sirtuin-1 on embryo implantation in oxidative stress–induced mice. J. Assist. Reprod. Genet. 2021, 38, 2349–2361. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Li, X.; He, Y.; Han, L.; Qiu, D.; Ling, L.; Liu, H.; Liu, J.; Gu, L. SIRT7 functions in redox homeostasis and cytoskeletal organization during oocyte maturation. FASEB J. 2018, 32, 6228–6238. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Liu, Y.; Rubio, V.; He, J.; Minze, L.J.; Shi, Z.-Z. OLA1, a Translational Regulator of p21, Maintains Optimal Cell Proliferation Necessary for Developmental Progression. Mol. Cell. Biol. 2016, 36, 2568–2582. [Google Scholar] [CrossRef] [Green Version]
- Ntostis, P.; Kokkali, G.; Iles, D.; Huntriss, J.; Tzetis, M.; Picton, H.; Pantos, K.; Miller, D. Can trophectoderm RNA analysis predict human blastocyst competency? Syst. Biol. Reprod. Med. 2019, 65, 312–325. [Google Scholar] [CrossRef] [Green Version]
- Hamatani, T.; Ko, M.S.; Yamada, M.; Kuji, N.; Mizusawa, Y.; Shoji, M.; Hada, T.; Asada, H.; Maruyama, T.; Yoshimura, Y. Global gene expression profiling of preimplantation embryos. Hum. Cell 2006, 19, 98–117. [Google Scholar] [CrossRef]
- Groff, A.F.; Resetkova, N.; DiDomenico, F.; Sakkas, D.; Penzias, A.; Rinn, J.L.; Eggan, K. RNA-seq as a tool for evaluating human embryo competence. Genome Res. 2019, 29, 1705–1718. [Google Scholar] [CrossRef] [Green Version]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Reyes, A.P.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Schultz, R.M. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 2005, 283, 40–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]

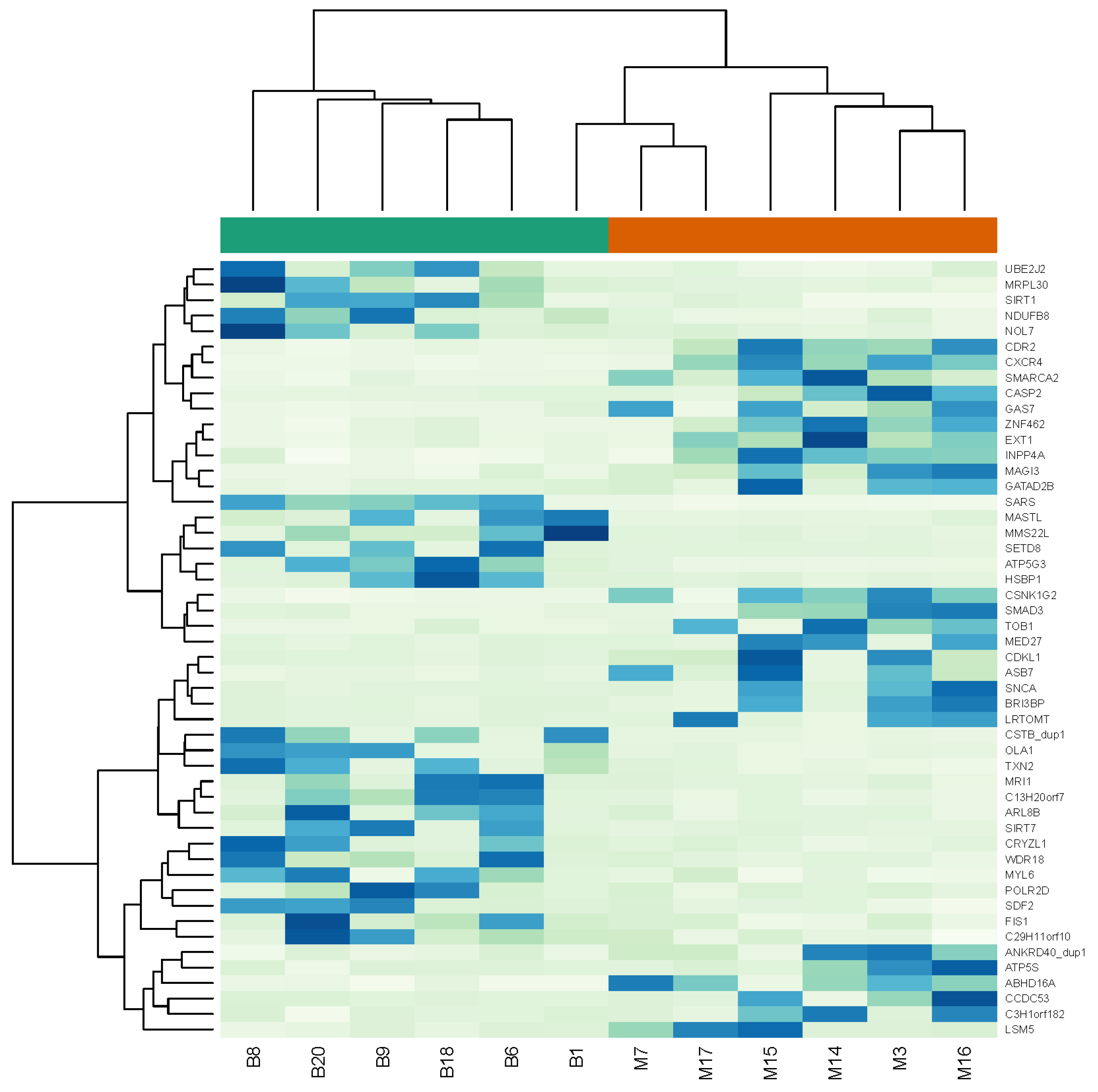
| Symbol | Gene Name | log2 Fold Change | p-Value |
|---|---|---|---|
| SARS | seryl-tRNA synthetase | −4.25 | 6.81 × 10−14 |
| HSBP1 | heat shock factor binding protein 1 | −3.87 | 3.42 × 10−7 |
| ATP5G3 | ATP synthase, H+ transporting, mitochondrial Fo complex subunit C3 (subunit 9) | −3.77 | 2.72 × 10−9 |
| C13H20orf7 | NA | −3.74 | 4.18 × 10−8 |
| CSTB_dup1 | NA | −3.65 | 3.83 × 10−8 |
| MRPL30 | mitochondrial ribosomal protein L30 | −3.61 | NA |
| MMS22L | MMS22 like, DNA repair protein | −3.57 | NA |
| MRI1 | methylthioribose-1-phosphate isomerase 1 | −3.56 | 1.04 × 10−6 |
| UCHL3_dup2 | NA | −3.55 | NA |
| NDUFB8 | NADH:ubiquinone oxidoreductase subunit B8 | −3.52 | 1.97 × 10−7 |
| Symbol | Gene Name | log2 Fold Change | p-Value |
|---|---|---|---|
| CDKL1 | cyclin dependent kinase like 1 | 3.31 | 4.37 × 10−6 |
| GATAD2B | GATA zinc finger domain containing 2B | 3.32 | 3.63 × 10−6 |
| LSM5 | LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated | 3.32 | 7.66 × 10−7 |
| C14H8orf59 | chromosome 14 open reading frame, human C8orf59 | 3.33 | NA |
| BRI3BP | BRI3 binding protein | 3.34 | 4.02 × 10−6 |
| MED27 | mediator complex subunit 27 | 3.37 | 3.03 × 10−6 |
| SNCA | synuclein α | 3.40 | 2.76 × 10−6 |
| CDR2 | cerebellar degeneration related protein 2 | 3.40 | 9.00 × 10−9 |
| EXT1 | exostosin glycosyltransferase 1 | 3.46 | NA |
| CXCR4 | chemokine (C-X-C motif) receptor 4 | 4.10 | 2.34 × 10−12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nõmm, M.; Ivask, M.; Pärn, P.; Reimann, E.; Kõks, S.; Jaakma, Ü. Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq. Genes 2023, 14, 569. https://doi.org/10.3390/genes14030569
Nõmm M, Ivask M, Pärn P, Reimann E, Kõks S, Jaakma Ü. Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq. Genes. 2023; 14(3):569. https://doi.org/10.3390/genes14030569
Chicago/Turabian StyleNõmm, Monika, Marilin Ivask, Pille Pärn, Ene Reimann, Sulev Kõks, and Ülle Jaakma. 2023. "Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq" Genes 14, no. 3: 569. https://doi.org/10.3390/genes14030569
APA StyleNõmm, M., Ivask, M., Pärn, P., Reimann, E., Kõks, S., & Jaakma, Ü. (2023). Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq. Genes, 14(3), 569. https://doi.org/10.3390/genes14030569





