Head Size in Phelan–McDermid Syndrome: A Literature Review and Pooled Analysis of 198 Patients Identifies Candidate Genes on 22q13
Abstract
1. Introduction
2. Materials and Methods
Statistical Analysis
3. Results
3.1. Investigation of Factors Associated with Head Size
3.2. Assessment of Deletion Location on Macrocephaly, Microcephaly, and Normocephaly
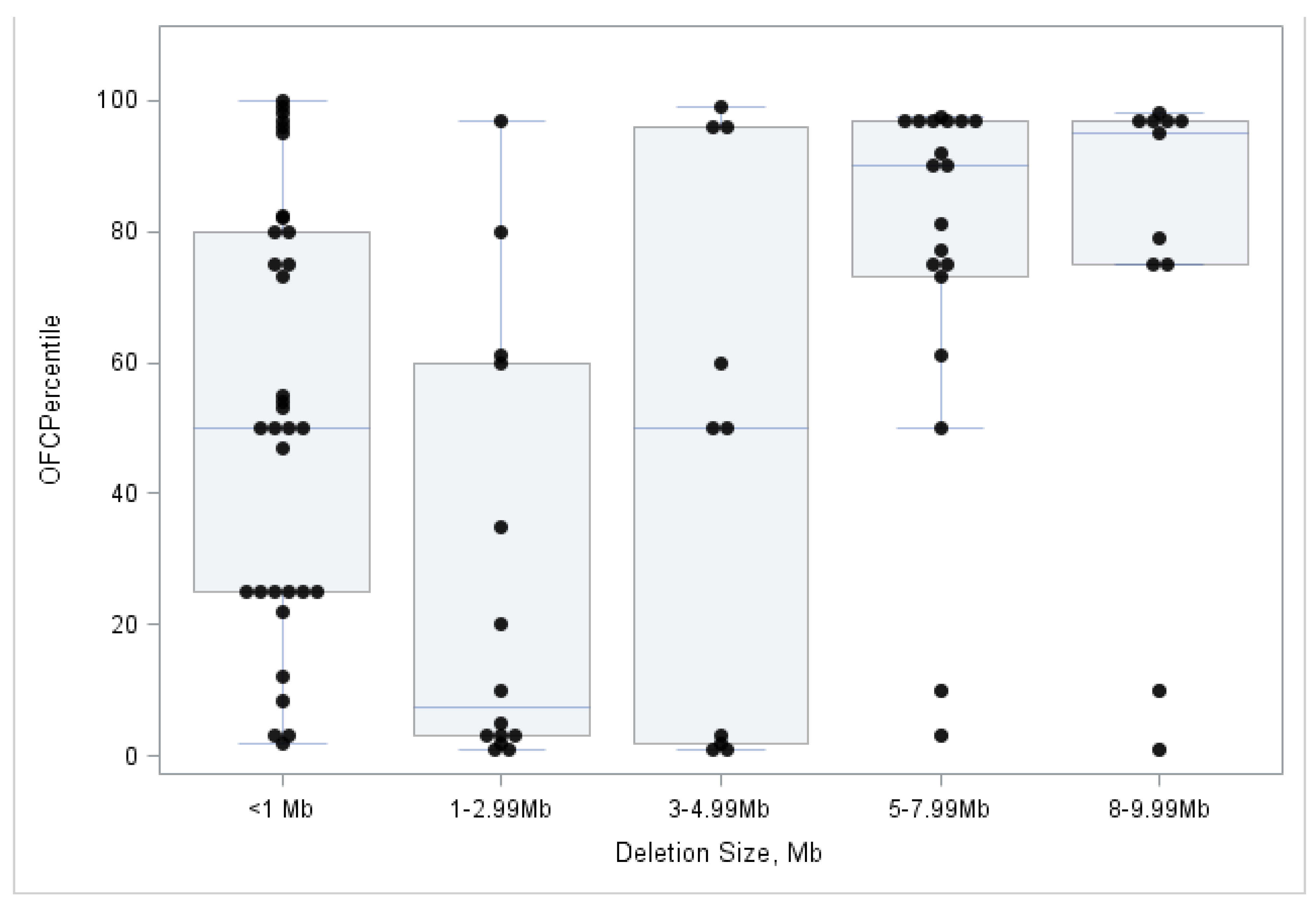
3.3. Association between OFC Measures and 22q13 Deletion Size
3.4. Association between Head Size and MRI Results
3.5. Association between 22q13 Genomic Deletions and Macrocephaly
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Phelan, K.; Rogers, R.C.; Boccuto, L. Phelan-McDermid Syndrome. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2018. [Google Scholar]
- Sarasua, S.M.; Boccuto, L.; Sharp, J.L.; Dwivedi, A.; Chen, C.; Rollins, J.D.; Rogers, R.C.; Phelan, K.; DuPont, B.R. Clinical and Genomic Evaluation of 201 Patients with Phelan-McDermid Syndrome. Hum. Genet. 2014, 133, 847–859. [Google Scholar] [CrossRef]
- Nevado, J.; García-Miñaúr, S.; Palomares-Bralo, M.; Vallespín, E.; Guillén-Navarro, E.; Rosell, J.; Bel-Fenellós, C.; Mori, M.Á.; Milá, M.; Del Campo, M.; et al. Variability in Phelan-McDermid Syndrome in a Cohort of 210 Individuals. Front. Genet. 2022, 13, 652454. [Google Scholar] [CrossRef]
- Rollins, J.D.; Sarasua, S.M.; Phelan, K.; DuPont, B.R.; Rogers, R.C.; Collins, J.S. Growth in Phelan-McDermid Syndrome. Am. J. Med. Genet. A 2011, 155A, 2324–2326. [Google Scholar] [CrossRef]
- Disciglio, V.; Lo Rizzo, C.; Mencarelli, M.A.; Mucciolo, M.; Marozza, A.; Di Marco, C.; Massarelli, A.; Canocchi, V.; Baldassarri, M.; Ndoni, E.; et al. Interstitial 22q13 Deletions Not Involving SHANK3 Gene: A New Contiguous Gene Syndrome. Am. J. Med. Genet. A 2014, 164A, 1666–1676. [Google Scholar] [CrossRef]
- Sarasua, S.M.; Dwivedi, A.; Boccuto, L.; Rollins, J.D.; Chen, C.; Rogers, R.C.; Phelan, K.; DuPont, B.R.; Collins, J.S. Association between Deletion Size and Important Phenotypes Expands the Genomic Region of Interest in Phelan-McDermid Syndrome (22q13 Deletion Syndrome). J. Med. Genet. 2011, 48, 761–766. [Google Scholar] [CrossRef]
- Jeffries, A.R.; Curran, S.; Elmslie, F.; Sharma, A.; Wenger, S.; Hummel, M.; Powell, J. Molecular and Phenotypic Characterization of Ring Chromosome 22. Am. J. Med. Genet. A 2005, 137, 139–147. [Google Scholar] [CrossRef]
- Bonaglia, M.C.; Giorda, R.; Beri, S.; De Agostini, C.; Novara, F.; Fichera, M.; Grillo, L.; Galesi, O.; Vetro, A.; Ciccone, R.; et al. Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011, 7, e1002173. [Google Scholar] [CrossRef]
- Phelan, K.; Boccuto, L.; Powell, C.M.; Boeckers, T.M.; van Ravenswaaij-Arts, C.; Rogers, R.C.; Sala, C.; Verpelli, C.; Thurm, A.; Bennett, W.E.; et al. Phelan-McDermid Syndrome: A Classification System After 30 years of Experience. Orphanet. J. Rare Dis. 2022, 17, 27. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Durand, C.M.; Betancur, C.; Boeckers, T.M.; Bockmann, J.; Chaste, P.; Fauchereau, F.; Nygren, G.; Rastam, M.; Gillberg, I.C.; Anckarsater, H.; et al. Mutations in the Gene Encoding the Synaptic Scaffolding Protein SHANK3 are Associated with Autism Spectrum Disorders. Nat. Genet. 2007, 39, 25–27. [Google Scholar] [CrossRef]
- Leblond, C.S.; Nava, C.; Polge, A.; Gauthier, J.; Huguet, G.; Lumbroso, S.; Giuliano, F.; Stordeur, C.; Depienne, C.; Mouzat, K.; et al. Meta-Analysis of SHANK Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments. PLoS Genet. 2014, 10, e1004580. [Google Scholar] [CrossRef] [PubMed]
- Moessner, R.; Marshall, C.R.; Sutcliffe, J.S.; Skaug, J.; Pinto, D.; Vincent, J.; Zwaigenbaum, L.; Fernandez, B.; Roberts, W.; Szatmari, P.; et al. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. Am. J. Hum. Genet. 2007, 81, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.P.; Griesi-Oliveira, K.; Bossolani-Martins, A.L.; Lourenço, N.C.V.; Takahashi, V.N.O.; da Rocha, K.M.; Moreira, E.S.; Vadasz, E.; Meira, J.G.C.; Bertola, D.; et al. Investigation of 15q11-q13, 16p11.2 and 22q13 CNVs in Autism Spectrum Disorder Brazilian Individuals with and without Epilepsy. PLoS ONE 2014, 9, e107705. [Google Scholar] [CrossRef]
- Guilmatre, A.; Huguet, G.; Delorme, R.; Bourgeron, T. The Emerging Role of SHANK Genes in Neuropsychiatric Disorders. Dev. Neurobiol. 2014, 74, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Anderlid, B.M.; Schoumans, J.; Anneren, G.; Tapia-Paez, I.; Dumanski, J.; Blennow, E.; Nordenskjold, M. FISH-Mapping of a 100-Kb Terminal 22q13 Deletion. Hum. Genet. 2002, 110, 439–443. [Google Scholar] [CrossRef]
- Bartsch, O.; Schneider, E.; Damatova, N.; Weis, R.; Tufano, M.; Iorio, R.; Ahmed, A.; Beyer, V.; Zechner, U.; Haaf, T. Fulminant Hepatic Failure Requiring Liver Transplantation in 22q13.3 Deletion Syndrome. Am. J. Med. Genet. A 2010, 152A, 2099–2102. [Google Scholar] [CrossRef]
- Battini, R.; Battaglia, A.; Bertini, V.; Cioni, G.; Parrini, B.; Rapalini, E.; Simi, P.; Tinelli, F.; Valetto, A. Characterization of the Phenotype and Definition of the Deletion in a New Patient with Ring Chromosome 22. Am. J. Med. Genet. A 2004, 130A, 196–199. [Google Scholar] [CrossRef]
- Bisgaard, A.-M.; Kirchhoff, M.; Nielsen, J.E.; Kibaek, M.; Lund, A.; Schwartz, M.; Christensen, E. Chromosomal Deletion Unmasking a Recessive Disease: 22q13 Deletion Syndrome and Metachromatic Leukodystrophy. Clin. Genet. 2009, 75, 175–179. [Google Scholar] [CrossRef]
- Boccuto, L.; Abenavoli, L.; Cascio, L.; Srikanth, S.; DuPont, B.; Mitz, A.R.; Rogers, R.C.; Phelan, K. Variability in Phelan-McDermid Syndrome: The Impact of the PNPLA3 P.I148M Polymorphism. Clin. Genet. 2018, 94, 590–591. [Google Scholar] [CrossRef]
- Bonaglia, M.C.; Giorda, R.; Mani, E.; Aceti, G.; Anderlid, B.-M.; Baroncini, A.; Pramparo, T.; Zuffardi, O. Identification of a Recurrent Breakpoint within the SHANK3 Gene in the 22q13.3 Deletion Syndrome. J. Med. Genet. 2006, 43, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Bonaglia, M.C.; Giorda, R.; Beri, S.; Bigoni, S.; Sensi, A.; Baroncini, A.; Capucci, A.; De Agostini, C.; Gwilliam, R.; Deloukas, P.; et al. Mosaic 22q13 Deletions: Evidence for Concurrent Mosaic Segmental Isodisomy and Gene Conversion. Eur. J. Hum. Genet. 2009, 17, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Breckpot, J.; Vercruyssen, M.; Weyts, E.; Vandevoort, S.; D’Haenens, G.; Van Buggenhout, G.; Leempoels, L.; Brischoux-Boucher, E.; Van Maldergem, L.; Renieri, A.; et al. Copy Number Variation Analysis in Adults with Catatonia Confirms Haploinsufficiency of SHANK3 as a Predisposing Factor. Eur. J. Med. Genet. 2016, 59, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lin, S.; Chern, S.; Tsai, F.; Wu, P.; Lee, C.; Chen, Y.; Chen, W.; Wang, W. A De Novo 7.9 Mb Deletion in 22q13.2→qter in a Boy with Autistic Features, Epilepsy, Developmental Delay, Atopic Dermatitis and Abnormal Immunological Findings. Eur. J. Med. Genet. 2010, 53, 329–332. [Google Scholar] [CrossRef]
- Cho, E.H.; Park, J.B.; Kim, J.K. Atypical Teratoid Rhabdoid Brain Tumor in an Infant with Ring Chromosome 22. Korean J. Pediatr. 2014, 57, 333–336. [Google Scholar] [CrossRef]
- Deibert, E.; Crenshaw, M.; Miller, M.S. A Patient with Phelan-McDermid Syndrome and Dilation of the Great Vessels. Clin. Case Rep. 2019, 7, 607–611. [Google Scholar] [CrossRef]
- Denayer, A.; Van Esch, H.; de Ravel, T.; Frijns, J.-P.; Van Buggenhout, G.; Vogels, A.; Devriendt, K.; Geutjens, J.; Thiry, P.; Swillen, A. Neuropsychopathology in 7 Patients with the 22q13 Deletion Syndrome: Presence of Bipolar Disorder and Progressive Loss of Skills. Mol. Syndromol. 2012, 3, 14–20. [Google Scholar] [CrossRef]
- Droogmans, G.; Swillen, A.; Van Buggenhout, G. Deep Phenotyping of Development, Communication and Behaviour in Phelan-McDermid Syndrome. Mol. Syndromol. 2020, 10, 294–305. [Google Scholar] [CrossRef]
- Figura, M.G.; Coppola, A.; Bottitta, M.; Calabrese, G.; Grillo, L.; Luciano, D.; Del Gaudio, L.; Torniero, C.; Striano, S.; Elia, M. Seizures and EEG Pattern in the 22q13.3 Deletion Syndrome: Clinical Report of Six Italian Cases. Seizure 2014, 23, 774–779. [Google Scholar] [CrossRef]
- Goizet, C.; Excoffier, E.; Taine, L.; Taupiac, E.; El Moneim, A.A.; Arveiler, B.; Bouvard, M.; Lacombe, D. Case with Autistic Syndrome and Chromosome 22q13.3 Deletion Detected by FISH. Am. J. Med. Genet. 2000, 96, 839–844. [Google Scholar] [CrossRef]
- Gong, X.; Jiang, Y.; Zhang, X.; An, Y.; Zhang, J.; Wu, Y.; Wang, J.; Sun, Y.; Liu, Y.; Gao, X.; et al. High Proportion of 22q13 Deletions and SHANK3 Mutations in Chinese Patients with Intellectual Disability. PLoS ONE 2012, 7, e34739. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, R.S.; Soares, K.C.; Simioni, M.; Vieira, T.P.; Gil-da-Silva-Lopes, V.L.; Kim, C.A.; Brunoni, D.; Spinner, N.B.; Conlin, L.K.; Christofolini, D.M.; et al. Clinical, Cytogenetic, and Molecular Characterization of Six Patients with Ring Chromosomes 22, Including One with Concomitant 22q11.2 Deletion. Am. J. Med. Genet. A 2014, 164A, 1659–1665. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.F.; Ahmad, A.; Lesperance, M.M. Clinical Characterization of Novel Chromosome 22q13 Microdeletions. Int. J. Pediatr. Otorhinolaryngol. 2016, 95, 121–126. [Google Scholar] [CrossRef]
- Hackmann, K.; Rump, A.; Haas, S.A.; Lemke, J.R.; Fryns, J.; Tzschach, A.; Wieczorek, D.; Albrecht, B.; Kuechler, A.; Ripperger, T.; et al. Tentative Clinical Diagnosis of Lujan-Fryns Syndrome—A Conglomeration of Different Genetic Entities? Am. J. Med. Genet. A 2016, 170A, 94–102. [Google Scholar] [CrossRef]
- Hannachi, H.; Mougou, S.; Benabdallah, I.; Soayh, N.; Kahloul, N.; Gaddour, N.; Le Lorc’h, M.; Sanlaville, D.; El Ghezal, H.; Saad, A. Molecular and Phenotypic Characterization of Ring Chromosome 22 in Two Unrelated Patients. Cytogenet. Genome Res. 2013, 140, 1–11. [Google Scholar] [CrossRef]
- Holder, J.L.; Quach, M.M. The Spectrum of Epilepsy and Electroencephalographic Abnormalities due to SHANK3 Loss-of-Function Mutations. Epilepsia 2016, 57, 1651–1659. [Google Scholar] [CrossRef]
- Ishikawa, N.; Kobayashi, Y.; Fujii, Y.; Yamamoto, T.; Kobayashi, M. Late-Onset Epileptic Spasms in a Patient with 22q13.3 Deletion Syndrome. Brain Dev. 2015, 38, 109–112. [Google Scholar] [CrossRef]
- Jungová, P.; Čumová, A.; Kramarová, V.; Lisyová, J.; Ďurina, P.; Chandoga, J.; Böhmer, D. Phelan-McDermid Syndrome in Adult Patient with Atypical Bipolar Psychosis Repeatedly Triggered by Febrility. Neurocase 2018, 24, 227–230. [Google Scholar] [CrossRef]
- Karaman, A.; Aydin, H.; Geçkinli, B.; Göksu, K. The Deletion 22q13 Syndrome: A New Case. Genet. Couns. 2015, 26, 53–60. [Google Scholar]
- Kirkpatrick, B.E.; El-Khechen, D. A Unique Presentation of 22q13 Deletion Syndrome: Multicystic Kidney, Orofacial Clefting, and Wilms’ Tumor. Clin. Dysmorphol. 2011, 20, 53–54. [Google Scholar] [CrossRef]
- Koolen, D.A.; Reardon, W.; Rosser, E.M.; Lacombe, D.; Hurst, J.A.; Law, C.J.; Bongers, E.M.H.F.; Ravenswaaij-Arts, C.M.A.v.; Leisink, M.A.R.; Geurts van Kessel, A.H.M.; et al. Molecular Characterisation of Patients with Subtelomeric 22q Abnormalities using Chromosome Specific Array-Based Comparative Genomic Hybridisation. Eur. J. Hum. Genet. EJHG 2005, 13, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Kurtas, N.; Arrigoni, F.; Errichiello, E.; Zucca, C.; Maghini, C.; D’Angelo, M.G.; Beri, S.; Giorda, R.; Bertuzzo, S.; Delledonne, M.; et al. Chromothripsis and Ring Chromosome 22: A Paradigm of Genomic Complexity in the Phelan-McDermid Syndrome (22q13 Deletion Syndrome). J. Med. Genet. 2018, 55, 269–277. [Google Scholar] [CrossRef]
- Lindquist, S.G.; Kirchhoff, M.; Lundsteen, C.; Pedersen, W.; Erichsen, G.; Kristensen, K.; Lillquist, K.; Smedegaard, H.H.; Skov, L.; Tommerup, N.; et al. Further Delineation of the 22q13 Deletion Syndrome. Clin. Dysmorphol. 2005, 14, 55–60. [Google Scholar] [CrossRef]
- Lumaka, A.; Race, V.; Peeters, H.; Corveleyn, A.; Coban-Akdemir, Z.; Jhangiani, S.N.; Song, X.; Mubungu, G.; Posey, J.; Lupski, J.R.; et al. A Comprehensive Clinical and Genetic Study in 127 Patients with ID in Kinshasa, DR Congo. Am. J. Med. Genet. A 2018, 176, 1897–1909. [Google Scholar] [CrossRef] [PubMed]
- Lund, C.; Brodtkorb, E.; Røsby, O.; Rødningen, O.K.; Selmer, K.K. Copy Number Variants in Adult Patients with Lennox–Gastaut Syndrome Features. Epilepsy Res. 2013, 105, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Macedoni-Lukšič, M.; Krgović, D.; Zagradišnik, B.; Kokalj-Vokač, N. Deletion of the Last Exon of SHANK3 Gene Produces the Full Phelan–McDermid Phenotype: A Case Report. Gene 2013, 524, 386–389. [Google Scholar] [CrossRef]
- Misceo, D.; Rødningen, O.K.; Barøy, T.; Sorte, H.; Mellembakken, J.R.; Strømme, P.; Fannemel, M.; Frengen, E. A Translocation between Xq21.33 and 22q13.33 Causes an Intragenic SHANK3 Deletion in a Woman with Phelan-McDermid Syndrome and Hypergonadotropic Hypogonadism. Am. J. Med. Genet. A 2011, 155A, 403–408. [Google Scholar] [CrossRef]
- Palumbo, P.; Accadia, M.; Leone, M.P.; Palladino, T.; Stallone, R.; Carella, M.; Palumbo, O. Clinical and Molecular Characterization of an Emerging Chromosome 22q13.31 Microdeletion Syndrome. Am. J. Med. Genet. Part A 2018, 176, 391–398. [Google Scholar] [CrossRef]
- Pan, L.; Sun, Y.; Chen, S.; He, J.; Xu, C. SNP Microarray Characterization and Genotype-Phenotype Analysis in a Patient with a Ring Chromosome 22. Int. J. Hum. Genet. 2014, 14, 23–26. [Google Scholar] [CrossRef]
- Philippe, A.; Boddaert, N.; Vaivre-Douret, L.; Robel, L.; Danon-Boileau, L.; Malan, V.; de Blois, M.; Heron, D.; Colleaux, L.; Golse, B.; et al. Neurobehavioral Profile and Brain Imaging Study of the 22q13.3 Deletion Syndrome in Childhood. Pediatrics 2008, 122, e376–e382. [Google Scholar] [CrossRef]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of Genes and Cellular Pathways Dysregulated in Autism Spectrum Disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Sathyamoorthi, S.; Morales, J.; Bermudez, J.; McBride, L.; Luquette, M.; McGoey, R.; Oates, N.; Hales, S.; Biegel, J.A.; Lacassie, Y. Array Analysis and Molecular Studies of INI1 in an Infant with Deletion 22q13 (Phelan–McDermid Syndrome) and Atypical Teratoid/Rhabdoid Tumor. Am. J. Med. Genet. Part A 2009, 149A, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Simenson, K.; Õiglane-Shlik, E.; Teek, R.; Kuuse, K.; Õunap, K. A Patient with the Classic Features of Phelan-McDermid Syndrome and a High Immunoglobulin E Level Caused by a Cryptic Interstitial 0.72-Mb Deletion in the 22q13.2 Region. Am. J. Med. Genet. A 2014, 164A, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Soorya, L.; Kolevzon, A.; Zweifach, J.; Lim, T.; Dobry, Y.; Schwartz, L.; Frank, Y.; Wang, A.T.; Cai, G.; Parkhomenko, E.; et al. Prospective Investigation of Autism and Genotype-Phenotype Correlations in 22q13 Deletion Syndrome and SHANK3 Deficiency. Mol. Autism 2013, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Tabet, A.; Rolland, T.; Ducloy, M.; Lévy, J.; Buratti, J.; Mathieu, A.; Haye, D.; Perrin, L.; Dupont, C.; Passemard, S.; et al. A Framework to Identify Contributing Genes in Patients with Phelan-McDermid Syndrome. NPJ Genom. Med. 2017, 2, 32. [Google Scholar] [CrossRef]
- Terrone, G.; Vitiello, G.; Genesio, R.; D’Amico, A.; Imperati, F.; Ugga, L.; Giugliano, T.; Piluso, G.; Nitsch, L.; Brunetti-Pierri, N.; et al. A Novel SHANK3 Interstitial Microdeletion in a Family with Intellectual Disability and Brain MRI Abnormalities Resembling Unidentified Bright Objects. Eur. J. Paediatr. Neurol. 2017, 21, 902–906. [Google Scholar] [CrossRef]
- Thümmler, S.; Giuliano, F.; Karmous-Benailly, H.; Richelme, C.; Fernandez, A.; De Georges, C.; Askenazy, F. Neurodevelopmental and Immunological Features in a Child Presenting 22q13.2 Microdeletion. Am. J. Med. Genet. A 2016, 170, 792–794. [Google Scholar] [CrossRef]
- Toruner, G.A.; Kurvathi, R.; Sugalski, R.; Shulman, L.; Twersky, S.; Pearson, P.G.; Tozzi, R.; Schwalb, M.N.; Wallerstein, R. Copy number variations in three children with sudden infant death. Clin. Genet. 2009, 76, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Upadia, J.; Gonzales, P.R.; Atkinson, T.P.; Schroeder, H.W.; Robin, N.H.; Rudy, N.L.; Mikhail, F.M. A Previously Unrecognized 22q13.2 Microdeletion Syndrome that Encompasses TCF20 and TNFRSF13C. Am. J. Med. Genet. A 2018, 176, 2791–2797. [Google Scholar] [CrossRef]
- Verhoeven, W.M.; Egger, J.I.; Willemsen, M.H.; de Leijer, G.J.; Kleefstra, T. Phelan-McDermid Syndrome in Two Adult Brothers: Atypical Bipolar Disorder as its Psychopathological Phenotype? Neuropsychiatr. Dis. Treat. 2012, 8, 175–179. [Google Scholar] [CrossRef]
- Verhoeven, W.M.A.; Egger, J.I.M.; Cohen-Snuijf, R.; Kant, S.G.; de Leeuw, N. Phelan-McDermid Syndrome: Clinical Report of a 70-Year-Old Woman. Am. J. Med. Genet. A 2013, 161A, 158–161. [Google Scholar] [CrossRef]
- Vlaskamp, D.R.M.; Callenbach, P.M.C.; Rump, P.; Giannini, L.A.A.; Dijkhuizen, T.; Brouwer, O.F.; van Ravenswaaij-Arts, C.M.A. Copy Number Variation in a Hospital-Based Cohort of Children with Epilepsy. Epilepsia Open 2017, 2, 244–254. [Google Scholar] [CrossRef]
- Vucurovic, K.; Landais, E.; Delahaigue, C.; Eutrope, J.; Schneider, A.; Leroy, C.; Kabbaj, H.; Motte, J.; Gaillard, D.; Rolland, A.; et al. Bipolar Affective Disorder and Early Dementia Onset in a Male Patient with SHANK3 Deletion. Eur. J. Med. Genet. 2012, 55, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, M.H.; Rensen, J.H.M.; van Schrojenstein-Lantman de Valk, H.M.J.; Hamel, B.C.J.; Kleefstra, T. Adult Phenotypes in Angelman- and Rett-Like Syndromes. Mol. Syndromol. 2012, 2, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ji, T.; Wang, J.; Xiao, J.; Wang, H.; Li, J.; Gao, Z.; Yang, Y.; Cai, B.; Wang, L.; et al. Submicroscopic Subtelomeric Aberrations in Chinese Patients with Unexplained Developmental Delay/Mental Retardation. BMC Med. Genet. 2010, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Lv, H.; Yang, T.; Du, X.; Sun, Y.; Xiao, B.; Fan, Y.; Luo, X.; Zhan, Y.; Wang, L.; et al. A 29 Mainland Chinese Cohort of Patients with Phelan–McDermid Syndrome: Genotype–phenotype Correlations and the Role of SHANK3 Haploinsufficiency in the Important Phenotypes. Orphanet. J. Rare Dis. 2020, 15, 335. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.L.; Crolla, J.A.; Walker, D.; Artifoni, L.; Dallapiccola, B.; Takano, T.; Vasudevan, P.; Huang, S.; Maloney, V.; Yobb, T.; et al. Interstitial 22q13 Deletions: Genes Other than have Major Effects on Cognitive and Language Development. Eur. J. Hum. Genet. EJHG 2008, 16, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Mitz, A.R.; Philyaw, T.J.; Boccuto, L.; Shcheglovitov, A.; Sarasua, S.M.; Kaufmann, W.E.; Thurm, A. Identification of 22q13 genes most likely to contribute to Phelan-McDermid Syndrome. Eur. J. Hum. Genet. 2018, 26, 293–302. [Google Scholar] [CrossRef]
- Ziegler, A.; Colin, E.; Goudenège, D.; Bonneau, D. A Snapshot of some pLI Score Pitfalls. Hum. Mutat. 2019, 40, 839–841. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Belhedi, N.; Bena, F.; Mrabet, A.; Guipponi, M.; Souissi, C.B.; Mrabet, H.K.; Elgaaied, A.B.; Malafosse, A.; Salzmann, A. A New Locus on Chromosome 22q13.31 Linked to Recessive Genetic Epilepsy with Febrile Seizures Plus (GEFS+) in a Tunisian Consanguineous Family. BMC Genet. 2013, 14, 93. [Google Scholar] [CrossRef]
- Jain, L.; Oberman, L.M.; Beamer, L.; Cascio, L.; May, M.; Srikanth, S.; Skinner, C.; Jones, K.; Allen, B.; Rogers, C.; et al. Genetic and Metabolic Profiling of individuals with Phelan-McDermid Syndrome Presenting with Seizures. Clin. Genet. 2022, 101, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Boucherie, C.; Boutin, C.; Jossin, Y.; Schakman, O.; Goffinet, A.M.; Ris, L.; Gailly, P.; Tissir, F. Neural Progenitor Fate Decision Defects, Cortical Hypoplasia and Behavioral Impairment in Celsr1-Deficient Mice. Mol. Psychiatry 2018, 23, 723–734. [Google Scholar] [CrossRef]
- Tenorio, J.; Nevado, J.; González-Meneses, A.; Arias, P.; Dapía, I.; Venegas-Vega, C.A.; Calvente, M.; Hernández, A.; Landera, L.; Ramos, S.; et al. Further Definition of the Proximal 19p13.3 Microdeletion/Microduplication Syndrome and Implication of PIAS4 as the Major Contributor. Clin. Genet. 2020, 97, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Sarasua, S.M.; Dwivedi, A.; Boccuto, L.; Chen, C.; Sharp, J.L.; Rollins, J.D.; Collins, J.S.; Rogers, R.C.; Phelan, K.; DuPont, B.R. 22q13.2q13.32 Genomic Regions Associated with Severity of Speech Delay, Developmental Delay, and Physical Features in Phelan-McDermid Syndrome. Genet. Med. 2014, 16, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Kanchwala, M.; Rios, J.J.; Hyatt, T.; Wang, R.C.; Tran, A.; Dougherty, I.; Tovar-Garza, A.; Purnadi, C.; Kumar, M.G.; et al. Biallelic Variants in RNU12 Cause CDAGS Syndrome. Hum. Mutat. 2021, 42, 1042–1052. [Google Scholar] [CrossRef]
- Malara, M.; Lutz, A.-K.; Incearap, B.; Bauer, H.F.; Cursano, S.; Volbracht, K.; Lerner, J.J.; Pandey, R.; Delling, J.P.; Ioannidis, V.; et al. SHANK3 deficiency leads to myelin defects in the central and peripheral nervous system. Cell. Mol. Life Sci. 2022, 79, 371. [Google Scholar] [CrossRef]
- Samogy-Costa, C.I.; Varella-Branco, E.; Monfardini, F.; Ferraz, H.; Fock, R.A.; Barbosa, R.H.A.; Pessoa, A.L.S.; Perez, A.B.A.; Lourenço, N.; Vibranovski, M.; et al. A Brazilian cohort of individuals with Phelan-McDermid syndrome: Genotype-phenotype correlation and identification of an atypical case. J. Neurodev. Disord. 2019, 11, 13. [Google Scholar] [CrossRef]
- Kothari, C.; Wack, M.; Hassen-Khodja, C.; Finan, S.; Savova, G.; O’Boyle, M.; Bliss, G.; Cornell, A.; Horn, E.J.; Davis, R.; et al. Phelan-McDermid syndrome data network: Integrating patient reported outcomes with clinical notes and curated genetic reports. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018, 177, 613–624. [Google Scholar] [CrossRef]


| Category | n (%) | |
|---|---|---|
| Year of Publication | 2000–2005 2006–2010 2011–2015 2016–2020 | 5 (9%) 11 (20%) 23 (41%) 17 (30%) |
| Country of Publication | Africa (DR Congo, Tunisia) Asia (China, Japan, Korea, Taiwan) Europe (Belgium, Czech Republic, Denmark, Estonia, France, Germany, Italy, Netherlands, Norway, Slovakia, Slovenia, Sweden, Turkey) North America (Canada, USA) South America (Brazil) | 2 (4%) 7 (13%) 34 (61%) 10 (18%) 3 (5%) |
| Sample Size with Data | 1 2–9 10–19 20–50 | 33 (59%) 19 (34%) 2 (4%) 2 (4%) |
| Variable | n = 198 |
|---|---|
| Sex Male Female | 93 (47%) 105 (53%) |
| Age of OFC measurement: Mean (SD) | 10.7 years (12.3) |
| Age of OFC measurement: n (%) Newborn—4 years 5–11 years 12–17 years 18–70 years | 82 (41%) 60 (30%) 20 (10%) 36 (18%) |
| Size of Deletion: mean (SD) | 3.34 Mb (2.96) |
| Size of Deletion: n (%) <100 kb 100–999 kb 1–2.99 Mb 3–4.99 Mb 5–6.99 Mb 7–9.4 Mb | 35 (18%) 28 (14%) 41 (21%) 29 (15%) 32 (16%) 33 (17%) |
| SHANK3 preserved: n (%) Yes No | 25 (13%) 173 (87%) |
| Mosaic: n (%) Yes No/unknown | 5 (3%) 193 (97%) |
| Inheritance: n (%) Maternally inherited Paternally inherited Inherited/unspecified De novo Unknown | 4 (2%) 6 (3%) 4 (4%) 115 (59%) 69 (35%) |
| Head size classification: n (%) Macrocephaly Normocephaly Microcephaly | 33 (17%) 139 (70%) 26 (13%) |
| OFC Percentile when reported: mean (SD) | 56.8 (SD 36.96) |
| OFC Percentile when reported: n (%) ≥97th 4th–96th ≤3rd missing | 26 (24%) 63 (58%) 19 (18%) 90 (--) |
| MRI Performed: n (%) Yes No | 97 (49%) 101 (51%) |
| MRI findings Normal Abnormal Type of abnormality * Atrophy Corpus callosum abnormalities Enlarged/abnormal ventricles Leukomalacia Cysts Hypomyelination Other | 26 (27%) 71 (73%) 12 (12%) 22 (22%) 21 (22%) 20 (21%) 12 (12%) 11 (11%) 11 (11%) |
| Macrocephaly | Normocephaly | Microcephaly | Kruskal Wallis p-Value | |
|---|---|---|---|---|
| n | n = 33 | n = 139 | n = 26 | |
| Median deletion size, Mb (hg19) | 5.32 Mb | 2.29 Mb | 2.20 Mb | 0.0310 |
| Median age | 6.0 | 5.25 | 6.0 | 0.5518 |
| % Male | 45% | 50% | 47% | 0.9377 |
| SHANK3 partially or fully deleted | n = 23 | n = 128 | n = 22 | |
| Median deletion size, Mb ± SD (hg19) | 5.62 (3.25) | 2.17 (3.07) | 2.29 (2.22) | 0.0788 |
| Median age in years (SD) | 7.0 (18.8) | 5.9 (11.6) | 5.0 (7.5) | 0.3776 |
| SHANK3 preserved | n = 10 | n = 11 | n = 4 | |
| Median deletion size, Mb ± SD (hg19) | 4.15 (1.61) | 4.60 (2.33) | 1.99 (3.30) | 0.6696 |
| Median age in years (SD) | 5.5 (6.89) | 7.0 (14.4) | 8.25 (6.25) | 0.7848 |
| Macrocephaly | Microcephaly | Normocephaly | Fisher’s Exact p-Value | |
|---|---|---|---|---|
| MRI (n = 97) Normal Abnormal | 3 (20%) 12 (80%) | 2 (14%) 12 (86%) | 21 (31%) 47 (69%) | 0.4322 |
| Brain Atrophy Yes No | 1 (7%) 14 (93%) | 5 (36%) 9 (64%) | 6 (9%) 62 (91%) | 0.0272 |
| Corpus Callosum ab. Yes No | 4 (27 %11 (73%) | 5 (36%) 9 (64%) | 13 (19%) 55 (81%) | 0.3354 |
| Cysts Yes No | 3 (20%) 12 (80%) | 0 (0%) 14 (100%) | 9 (13%) 68 (87%) | 0.2761 |
| Hypomyelination Yes No | 1 (7%) 14 (93% | 1 (7%) 13 (93%) | 9 (13%) 59 (87%) | 0.8908 |
| Leukomalacia Yes No | 5 (33%) 10 (67%) | 2 (14%) 12 (86%) | 13 (19%) 55 (81%) | 0.4594 |
| Enlarged/Abnormal ventricles Yes No | 4 (27%) 11 (73%) | 4 (29%) 10 (71%) | 13 (19%) 55 (81%) | 0.5468 |
| Other Abnormality Yes No | 2 (13%) 13 (87%) | 2 (14%) 12 (86%) | 7 (10%) 61 (90%) | 0.6853 |
| Head Size | Genomic Region Nominally Associated p < 0.05 | The Genomic Region Associated after Benjamini–Hochberg Correction (p < 0.05) | Genes Significant in Regional Tests after Benjamini–Hochberg Correction, p < 0.05 |
|---|---|---|---|
| Macrocephaly, SHANK3 preserved | 42.90–44.85 | 42.92–43.36 | 14 genes: SERHL2, RRP7B, POLDIP3, RNU12, CYB5R3, ATP5L2, A4GALT, ARFGAP3, PACSIN2, TTLL1, BIK, MCAT, TSPO, TTLL12 |
| Microcephaly, SHANK3 preserved | None | None | None |
| Macrocephaly, SHANK3 partially or fully deleted | 45.58–49.00 | 46.69–46.91 | 22 genes: ATXN10, WNT7B, LOC730668, LINC00899, PRR34, LOC150381, MIRLET7BHG, MIR3619, MIRLET7A3, MIR4763, MIRLET7B, PPARA, CDPF1, PKDREJ, TTC38, GTSE1-AS1, GTSE1, TRMU, CELSR1, GRAMD4, CERK, TBC1D22A |
| Microcephaly, SHANK3 partially or fully deleted | 50.00–50.94 | None | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarasua, S.M.; DeLuca, J.M.; Rogers, C.; Phelan, K.; Rennert, L.; Powder, K.E.; Weisensee, K.; Boccuto, L. Head Size in Phelan–McDermid Syndrome: A Literature Review and Pooled Analysis of 198 Patients Identifies Candidate Genes on 22q13. Genes 2023, 14, 540. https://doi.org/10.3390/genes14030540
Sarasua SM, DeLuca JM, Rogers C, Phelan K, Rennert L, Powder KE, Weisensee K, Boccuto L. Head Size in Phelan–McDermid Syndrome: A Literature Review and Pooled Analysis of 198 Patients Identifies Candidate Genes on 22q13. Genes. 2023; 14(3):540. https://doi.org/10.3390/genes14030540
Chicago/Turabian StyleSarasua, Sara M., Jane M. DeLuca, Curtis Rogers, Katy Phelan, Lior Rennert, Kara E. Powder, Katherine Weisensee, and Luigi Boccuto. 2023. "Head Size in Phelan–McDermid Syndrome: A Literature Review and Pooled Analysis of 198 Patients Identifies Candidate Genes on 22q13" Genes 14, no. 3: 540. https://doi.org/10.3390/genes14030540
APA StyleSarasua, S. M., DeLuca, J. M., Rogers, C., Phelan, K., Rennert, L., Powder, K. E., Weisensee, K., & Boccuto, L. (2023). Head Size in Phelan–McDermid Syndrome: A Literature Review and Pooled Analysis of 198 Patients Identifies Candidate Genes on 22q13. Genes, 14(3), 540. https://doi.org/10.3390/genes14030540






