The Prognostic Impact of Gender, Therapeutic Strategies, Molecular Background, and Tumor-Infiltrating Lymphocytes in Glioblastoma: A Still Unsolved Jigsaw
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Tissue Microarray
2.3. Immunohistochemistry and Imaging Analysis
2.4. Molecular Characterization
2.5. Statistical Analyses
3. Results
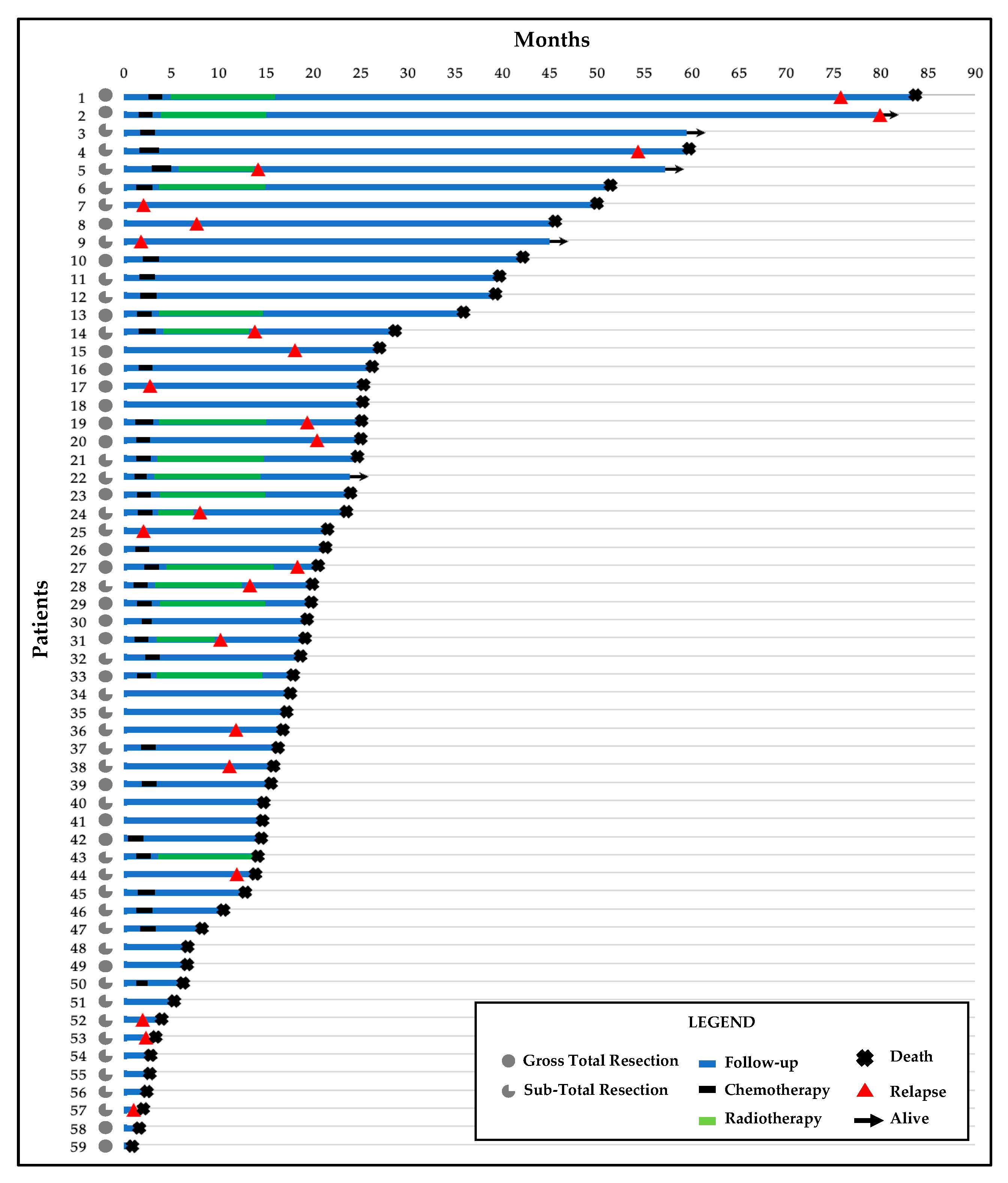
3.1. Clinicopathological Findings
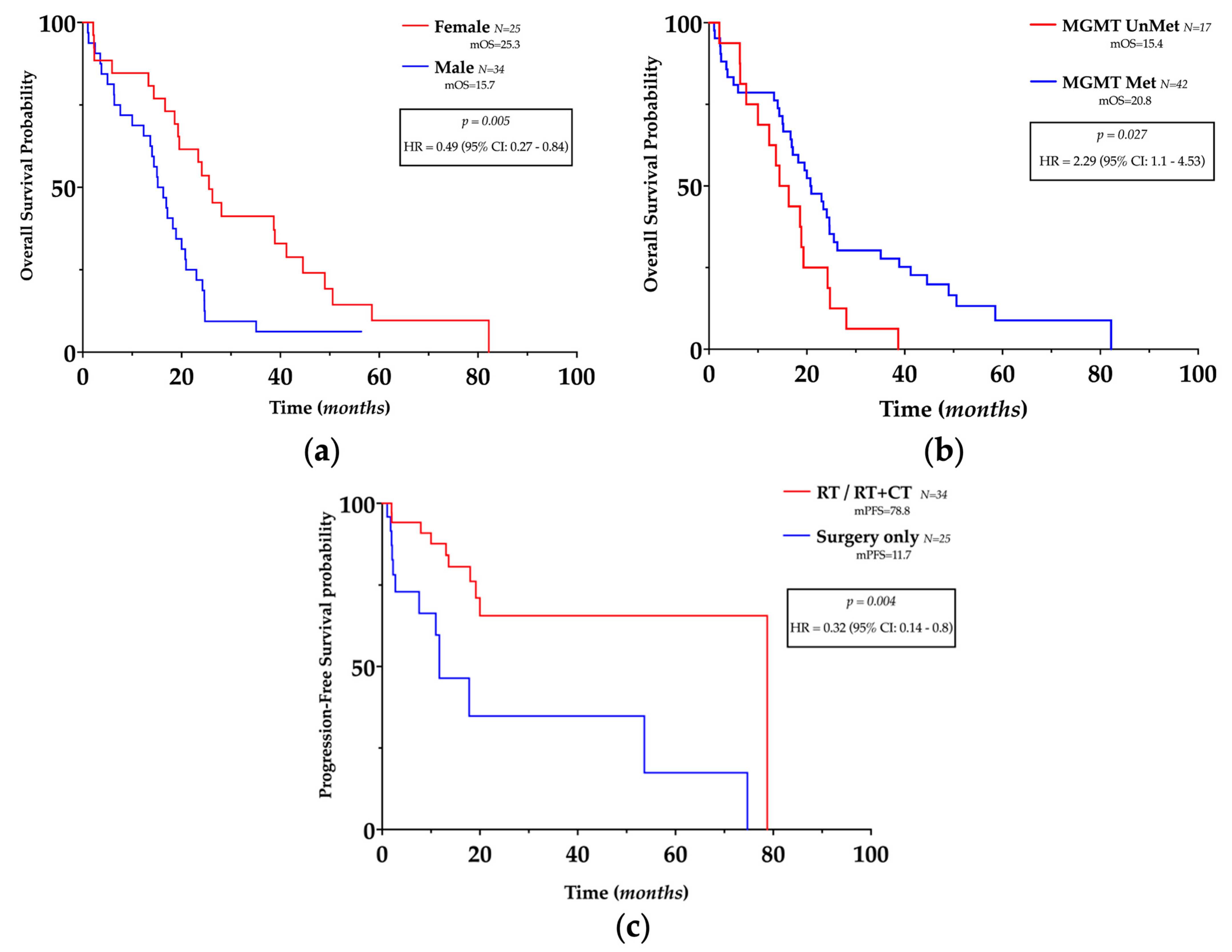
3.2. Clinico-Pathological Features and Prognosis
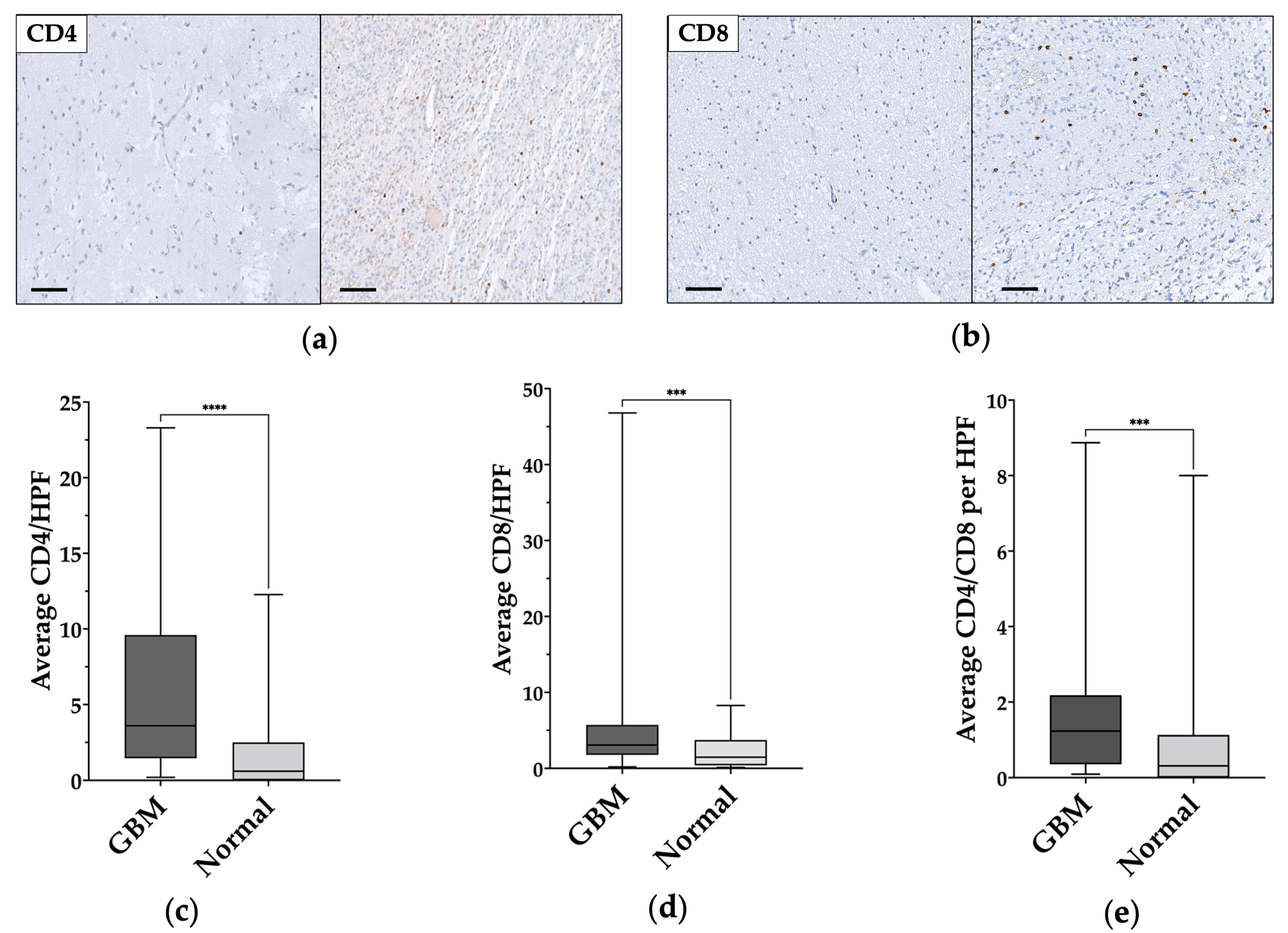
3.3. TILs Characterization in GBM and Normal Tissue
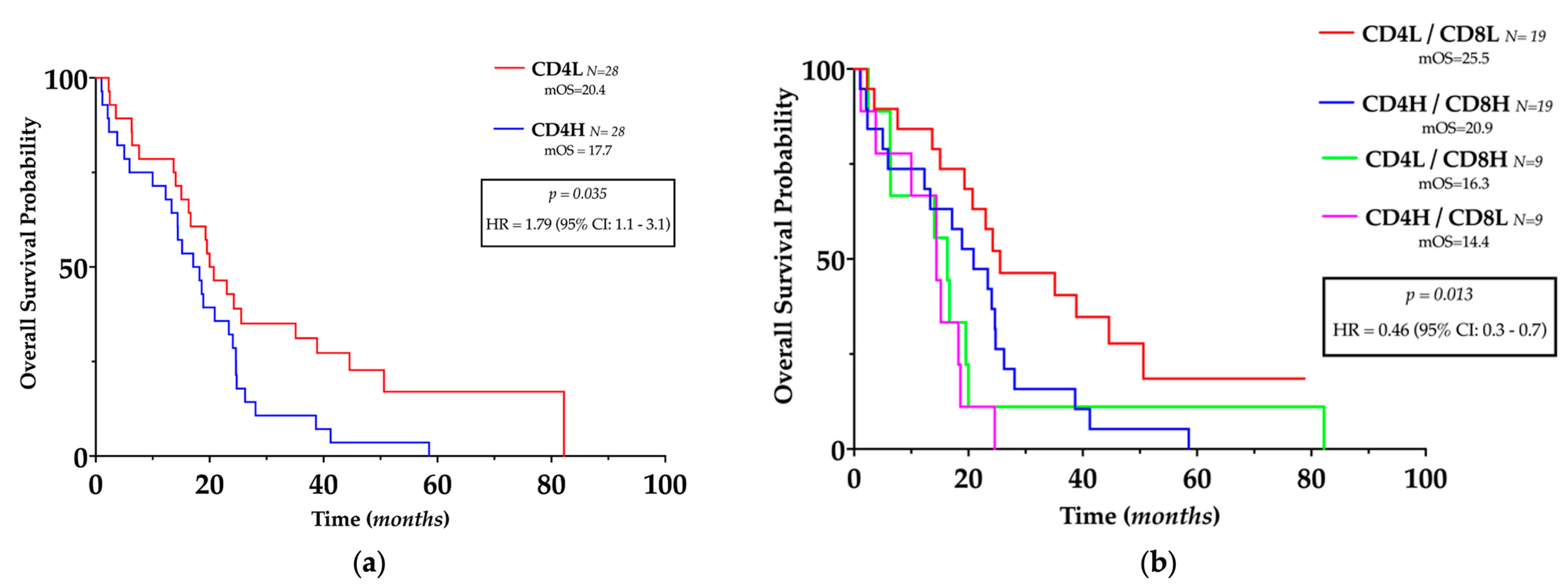
3.4. TILs Levels and Clinical Outcomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fanelli, G.N.; Grassini, D.; Ortenzi, V.; Pasqualetti, F.; Montemurro, N.; Perrini, P.; Naccarato, A.G.; Scatena, C. Decipher the Glioblastoma Microenvironment: The First Milestone for New Groundbreaking Therapeutic Strategies. Genes 2021, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wang, M.; Chen, Y.; Gong, J.; Chen, L.; Shi, X.; Lan, F.; Chen, Z.; Xiong, T.; Sun, H.; et al. Trends in Intracranial Glioma Incidence and Mortality in the United States, 1975–2018. Front Oncol. 2021, 11, 748061. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, W.C.; Kong, J.; Gao, J.; Gutman, D.A.; Cooper, L.A.; Appin, C.; Park, Y.; Scarpace, L.; Mikkelsen, T.; Cohen, M.L.; et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin. Cancer Res. 2013, 19, 4951–4960. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Weller, M.; Stupp, R.; Hegi, M.E.; van den Bent, M.; Tonn, J.C.; Sanson, M.; Wick, W.; Reifenberger, G. Personalized care in neuro-oncology coming of age: Why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012, 14 (Suppl. 4), iv100–iv108. [Google Scholar] [CrossRef]
- Khan, M.T.; Prajapati, B.; Lakhina, S.; Sharma, M.; Prajapati, S.; Chosdol, K.; Sinha, S. Identification of Gender-Specific Molecular Differences in Glioblastoma (GBM) and Low-Grade Glioma (LGG) by the Analysis of Large Transcriptomic and Epigenomic Datasets. Front Oncol. 2021, 11, 699594. [Google Scholar] [CrossRef]
- Tian, M.; Ma, W.; Chen, Y.; Yu, Y.; Zhu, D.; Shi, J.; Zhang, Y. Impact of gender on the survival of patients with glioblastoma. Biosci. Rep. 2018, 38, 752. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Naccarato, A.G.; Scatena, C. Recent Advances in Cancer Plasticity: Cellular Mechanisms, Surveillance Strategies, and Therapeutic Optimization. Front Oncol. 2020, 10, 569. [Google Scholar] [CrossRef]
- Turtoi, A.; Musmeci, D.; Naccarato, A.G.; Scatena, C.; Ortenzi, V.; Kiss, R.; Murtas, D.; Patsos, G.; Mazzucchelli, G.; De Pauw, E.; et al. Sparc-like protein 1 is a new marker of human glioma progression. J. Proteome Res. 2012, 11, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, B. The blood-central nervous system barriers actively control immune cell entry into the central nervous system. Curr. Pharm. Des. 2008, 14, 1555–1565. [Google Scholar] [CrossRef]
- Abels, E.R.; Maas, S.L.N.; Tai, E.; Ting, D.T.; Broekman, M.L.D.; Breakefield, X.O.; El Khoury, J. GlioM&M: Web-based tool for studying circulating and infiltrating monocytes and macrophages in glioma. Sci. Rep. 2020, 10, 9898. [Google Scholar] [CrossRef] [PubMed]
- Perus, L.J.M.; Walsh, L.A. Microenvironmental Heterogeneity in Brain Malignancies. Front. Immunol. 2019, 10, 2294. [Google Scholar] [CrossRef] [PubMed]
- Safdari, H.; Hochberg, F.H.; Richardson, E.P., Jr. Prognostic value of round cell (lymphocyte) infiltration in malignant gliomas. Surg. Neurol. 1985, 23, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, C.; Li, Q.; Dong, J.; Liu, Y.; Huang, Y.; Jiang, T.; Wu, A. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br. J. Cancer 2014, 110, 2560–2568. [Google Scholar] [CrossRef]
- Mahmoud, S.M.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.; Ellis, I.O.; Green, A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef]
- Rossi, M.L.; Jones, N.R.; Candy, E.; Nicoll, J.A.; Compton, J.S.; Hughes, J.T.; Esiri, M.M.; Moss, T.H.; Cruz-Sanchez, F.F.; Coakham, H.B. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol. 1989, 78, 189–193. [Google Scholar] [CrossRef]
- Schemper, M.; Smith, T.L. A note on quantifying follow-up in studies of failure time. Control. Clin. Trials 1996, 17, 343–346. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Scarpitta, R.; Cinacchi, P.; Fuochi, B.; Szumera-Cieckiewicz, A.; De Ieso, K.; Ferrari, P.; Fontana, A.; Miccoli, M.; Naccarato, A.G.; et al. Immunohistochemistry for Thymidine Kinase-1 (TK1): A Potential Tool for the Prognostic Stratification of Breast Cancer Patients. J. Clin. Med. 2021, 10, 5416. [Google Scholar] [CrossRef]
- Saraggi, D.; Galuppini, F.; Fanelli, G.N.; Remo, A.; Urso, E.D.L.; Bao, R.Q.; Bacchin, D.; Guzzardo, V.; Luchini, C.; Braconi, C.; et al. MiR-21 up-regulation in ampullary adenocarcinoma and its pre-invasive lesions. Pathol. Res. Pract. 2018, 214, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.N.; Gasparini, P.; Coati, I.; Cui, R.; Pakula, H.; Chowdhury, B.; Valeri, N.; Loupakis, F.; Kupcinskas, J.; Cappellesso, R.; et al. LONG-NONCODING RNAs in gastroesophageal cancers. Noncoding RNA Res. 2018, 3, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Facchin, S.; Munari, G.; Fanelli, G.N.; Lorenzon, G.; Savarino, E. Noncoding RNAs as drivers of the phenotypic plasticity of oesophageal mucosa. World J. Gastroenterol. 2017, 23, 7653–7656. [Google Scholar] [CrossRef] [PubMed]
- Saraggi, D.; Galuppini, F.; Remo, A.; Urso, E.D.L.; Bacchin, D.; Salmaso, R.; Lanza, C.; Bao, R.Q.; Fanelli, G.N.; Guzzardo, V.; et al. PD-L1 overexpression in ampulla of Vater carcinoma and its pre-invasive lesions. Histopathology 2017, 71, 470–474. [Google Scholar] [CrossRef]
- Fusco, N.; Ragazzi, M.; Sajjadi, E.; Venetis, K.; Piciotti, R.; Morganti, S.; Santandrea, G.; Fanelli, G.N.; Despini, L.; Invernizzi, M.; et al. Assessment of estrogen receptor low positive status in breast cancer: Implications for pathologists and oncologists. Histol. Histopathol. 2021, 36, 1235–1245. [Google Scholar] [CrossRef]
- Scatena, C.; Fanelli, G.; Fanelli, G.N.; Menicagli, M.; Aretini, P.; Ortenzi, V.; Civitelli, S.P.; Innocenti, L.; Sotgia, F.; Lisanti, M.P.; et al. New insights in the expression of stromal caveolin 1 in breast cancer spread to axillary lymph nodes. Sci. Rep. 2021, 11, 2755. [Google Scholar] [CrossRef]
- Forooshani, M.K.; Scarpitta, R.; Fanelli, G.N.; Miccoli, M.; Naccarato, A.G.; Scatena, C. Is It Time to Consider the Androgen Receptor as a Therapeutic Target in Breast Cancer? Anticancer Agents Med. Chem. 2022, 22, 775–786. [Google Scholar] [CrossRef]
- Francini, E.; Fanelli, G.N.; Pederzoli, F.; Spisak, S.; Minonne, E.; Raffo, M.; Pakula, H.; Tisza, V.; Scatena, C.; Naccarato, A.G.; et al. Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine. Cancers 2022, 14, 4359. [Google Scholar] [CrossRef]
- Penney, K.L.; Tyekucheva, S.; Rosenthal, J.; El Fandy, H.; Carelli, R.; Borgstein, S.; Zadra, G.; Fanelli, G.N.; Stefanizzi, L.; Giunchi, F.; et al. Metabolomics of Prostate Cancer Gleason Score in Tumor Tissue and Serum. Mol. Cancer Res. 2021, 19, 475–484. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Giampietro, C.; Montemurro, N.; Giannini, N.; Gadducci, G.; Orlandi, P.; Natali, E.; Chiarugi, P.; Gonnelli, A.; Cantarella, M.; et al. Old and New Systemic Immune-Inflammation Indexes Are Associated with Overall Survival of Glioblastoma Patients Treated with Radio-Chemotherapy. Genes 2022, 13, 1054. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Vianello, L.; Sacchi, D.; Fanelli, G.N.; Munari, G.; Scarpa, M.; Cappellesso, R.; Loupakis, F.; Lanza, C.; Salmaso, R.; et al. Assessment of intratumor immune-microenvironment in colorectal cancers with extranodal extension of nodal metastases. Cancer Cell Int. 2018, 18, 131, Erratum in Cancer Cell Int. 2019, 19, 244. [Google Scholar] [CrossRef]
- Claus, E.B.; Black, P.M. Survival rates and patterns of care for patients diagnosed with supratentorial low-grade gliomas: Data from the SEER program, 1973-2001. Cancer 2006, 106, 1358–1363. [Google Scholar] [CrossRef] [PubMed]
- Barone, T.A.; Gorski, J.W.; Greenberg, S.J.; Plunkett, R.J. Estrogen increases survival in an orthotopic model of glioblastoma. J. Neurooncol. 2009, 95, 37–48. [Google Scholar] [CrossRef]
- Li, Q.; Jedlicka, A.; Ahuja, N.; Gibbons, M.C.; Baylin, S.B.; Burger, P.C.; Issa, J.P. Concordant methylation of the ER and N33 genes in glioblastoma multiforme. Oncogene 1998, 16, 3197–3202. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, Y.; Wei, W.; Cong, P.; Ding, Y.; Xiang, L.; Wu, K. Androgen receptor signaling regulates growth of glioblastoma multiforme in men. Tumour Biol. 2015, 36, 967–972. [Google Scholar] [CrossRef]
- Weller, M.; Le Rhun, E. How did lomustine become standard of care in recurrent glioblastoma? Cancer Treat. Rev. 2020, 87, 102029. [Google Scholar] [CrossRef]
- Davis, F.G.; Freels, S.; Grutsch, J.; Barlas, S.; Brem, S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: An analysis based on Surveillance, Epidemiology, and End Results (SEER) data, 1973–1991. J. Neurosurg. 1998, 88, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.S.; Huang, F.P.; Zheng, K.; Zhang, H.S.; Zhou, X.; Bao, X.H.; Zheng, J.J.; Chang, C.; Zhou, L.F. Factors affecting prognosis of patients with intracranial anaplastic oligodendrogliomas: A single institutional review of 70 patients. J. Neurooncol. 2010, 100, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Li, X.; Yan, Y.; Shen, X.; Wang, J.; Yang, H.; Liu, S.; Han, C.; Hu, Y. Exploring the relationship between age and prognosis in glioma: Rethinking current age stratification. BMC Neurol. 2022, 22, 350. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Liao, J.; Long, Y. Comparative assessment of the efficacy of gross total versus subtotal total resection in patients with glioma: A meta-analysis. Int. J. Surg. 2019, 63, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Kramm, C.M.; Wagner, S.; Van Gool, S.; Schmid, H.; Strater, R.; Gnekow, A.; Rutkowski, S.; Wolff, J.E. Improved survival after gross total resection of malignant gliomas in pediatric patients from the HIT-GBM studies. Anticancer Res. 2006, 26, 3773–3779. [Google Scholar]
- Nam, J.Y.; de Groot, J.F. Treatment of Glioblastoma. J. Oncol. Pract. 2017, 13, 629–638. [Google Scholar] [CrossRef]
- Trifiletti, D.M.; Alonso, C.; Grover, S.; Fadul, C.E.; Sheehan, J.P.; Showalter, T.N. Prognostic Implications of Extent of Resection in Glioblastoma: Analysis from a Large Database. World Neurosurg. 2017, 103, 330–340. [Google Scholar] [CrossRef]
- Shonka, N.A.; Aizenberg, M.R. Extent of Resection in Glioblastoma. J. Oncol. Pract. 2017, 13, 641–642. [Google Scholar] [CrossRef]
- Li, Y.M.; Suki, D.; Hess, K.; Sawaya, R. The influence of maximum safe resection of glioblastoma on survival in 1229 patients: Can we do better than gross-total resection? J. Neurosurg. 2016, 124, 977–988. [Google Scholar] [CrossRef]
- Gessler, F.; Bernstock, J.D.; Braczynski, A.; Lescher, S.; Baumgarten, P.; Harter, P.N.; Mittelbronn, M.; Wu, T.; Seifert, V.; Senft, C. Surgery for Glioblastoma in Light of Molecular Markers: Impact of Resection and MGMT Promoter Methylation in Newly Diagnosed IDH-1 Wild-Type Glioblastomas. Neurosurgery 2019, 84, 190–197. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Montemurro, N.; Desideri, I.; Loi, M.; Giannini, N.; Gadducci, G.; Malfatti, G.; Cantarella, M.; Gonnelli, A.; Montrone, S.; et al. Impact of recurrence pattern in patients undergoing a second surgery for recurrent glioblastoma. Acta Neurol. Belg. 2022, 122, 441–446. [Google Scholar] [CrossRef]
- Montemurro, N.; Fanelli, G.N.; Scatena, C.; Ortenzi, V.; Pasqualetti, F.; Mazzanti, C.M.; Morganti, R.; Paiar, F.; Naccarato, A.G.; Perrini, P. Surgical outcome and molecular pattern characterization of recurrent glioblastoma multiforme: A single-center retrospective series. Clin. Neurol. Neurosurg. 2021, 207, 106735. [Google Scholar] [CrossRef]
- Indraccolo, S.; Lombardi, G.; Fassan, M.; Pasqualini, L.; Giunco, S.; Marcato, R.; Gasparini, A.; Candiotto, C.; Nalio, S.; Fiduccia, P.; et al. Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin. Cancer Res. 2019, 25, 1828–1837. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef]
- Brooks, W.H.; Markesbery, W.R.; Gupta, G.D.; Roszman, T.L. Relationship of lymphocyte invasion and survival of brain tumor patients. Ann. Neurol. 1978, 4, 219–224. [Google Scholar] [CrossRef]
- Palma, L.; Di Lorenzo, N.; Guidetti, B. Lymphocytic infiltrates in primary glioblastomas and recidivous gliomas. Incidence, fate, and relevance to prognosis in 228 operated cases. J. Neurosurg. 1978, 49, 854–861. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Dunn, G.P.; Schreiber, R.D. Cancer Immunosurveillance and Immunoediting: The Roles of Immunity in Suppressing Tumor Development and Shaping Tumor Immunogenicity. Adv. Immunol. 2006, 90, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Sarkaria, J.N.; Hu, L.S.; Parney, I.F.; Pafundi, D.H.; Brinkmann, D.H.; Laack, N.N.; Giannini, C.; Burns, T.C.; Kizilbash, S.H.; Laramy, J.K.; et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018, 20, 184–191. [Google Scholar] [CrossRef]
- Kmiecik, J.; Poli, A.; Brons, N.H.; Waha, A.; Eide, G.E.; Enger, P.O.; Zimmer, J.; Chekenya, M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013, 264, 71–83. [Google Scholar] [CrossRef]
- Waziri, A.; Killory, B.; Ogden, A.T., 3rd; Canoll, P.; Anderson, R.C.; Kent, S.C.; Anderson, D.E.; Bruce, J.N. Preferential in situ CD4+CD56+ T cell activation and expansion within human glioblastoma. J. Immunol. 2008, 180, 7673–7680. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Pages, F.; Kirilovsky, A.; Mlecnik, B.; Asslaber, M.; Tosolini, M.; Bindea, G.; Lagorce, C.; Wind, P.; Marliot, F.; Bruneval, P.; et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 2009, 27, 5944–5951. [Google Scholar] [CrossRef]
- Mauldin, I.S.; Jo, J.; Wages, N.A.; Yogendran, L.V.; Mahmutovic, A.; Young, S.J.; Lopes, M.B.; Slingluff, C.L., Jr.; Erickson, L.D.; Fadul, C.E. Proliferating CD8(+) T Cell Infiltrates Are Associated with Improved Survival in Glioblastoma. Cells 2021, 10, 3378. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Jung, T.Y.; Jung, S.; Jang, W.Y.; Moon, K.S.; Kim, I.Y.; Lee, M.C.; Lee, J.J. Tumour-infiltrating T-cell subpopulations in glioblastomas. Br. J. Neurosurg. 2012, 26, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Perrin, G.; Schnuriger, V.; Quiquerez, A.L.; Saas, P.; Pannetier, C.; de Tribolet, N.; Tiercy, J.M.; Aubry, J.P.; Dietrich, P.Y.; Walker, P.R. Astrocytoma infiltrating lymphocytes include major T cell clonal expansions confined to the CD8 subset. Int. Immunol. 1999, 11, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; DeNardo, D.G.; Affara, N.I.; Coussens, L.M. Lymphocytes in cancer development: Polarization towards pro-tumor immunity. Cytokine Growth Factor Rev. 2010, 21, 3–10. [Google Scholar] [CrossRef]
- Zamarron, B.F.; Chen, W. Dual roles of immune cells and their factors in cancer development and progression. Int. J. Biol. Sci. 2011, 7, 651–658. [Google Scholar] [CrossRef]
- Bos, R.; Marquardt, K.L.; Cheung, J.; Sherman, L.A. Functional differences between low- and high-affinity CD8(+) T cells in the tumor environment. Oncoimmunology 2012, 1, 1239–1247. [Google Scholar] [CrossRef]
- Bos, R.; Sherman, L.A. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 2010, 70, 8368–8377. [Google Scholar] [CrossRef]
- Mitsdoerffer, M.; Aly, L.; Barz, M.; Engleitner, T.; Sie, C.; Delbridge, C.; Lepennetier, G.; Ollinger, R.; Pfaller, M.; Wiestler, B.; et al. The glioblastoma multiforme tumor site promotes the commitment of tumor-infiltrating lymphocytes to the T(H)17 lineage in humans. Proc. Natl. Acad. Sci. USA 2022, 119, e2206208119. [Google Scholar] [CrossRef] [PubMed]
| Clinicopathological Data | |
|---|---|
| Number of Patients | 59 |
| Mean Age (range) | 62.15 y (26–80) |
| Gender | M = 34 (58%) |
| F = 25 (42%) | |
| Tumor site | Frontal = 12 (20%) |
| Parietal = 11 (19%) | |
| Temporal = 9 (15%) | |
| Occipital = 1 (2%) | |
| Fronto-parietal = 8 (13.5%) | |
| Fronto-temporal = 5 (8%) | |
| Temporo-parietal = 8 (13.5%) | |
| Parieto-occipital = 2 (3%) | |
| Temporo-occipital = 2 (3%) | |
| Insular 1 (3%) | |
| Extent of Surgical Resection | GTR = 24 (40%) |
| STR = 35 (60%) | |
| MGMT promoter | Met = 42 (71%) |
| Unmet = 17 (29%) | |
| IDH1 | Mut = 3 (5%) |
| WT = 56 (95%) | |
| IDH2 | Mut = 0 (0%) |
| WT = 59 (100%) | |
| 1p/19q co-deletion | Y = 10 (17%) |
| N = 49 (83%) | |
| Mean Proliferation Index (ki-67—range) | 33,4% (10–90%) |
| TP53 | Mut = 8 (13.5%) |
| WT = 51 (86.5%) | |
| Treatment | RT only = 16 (27%) |
| RT + CT = 18 (31%) | |
| N = 25 (42%) | |
| Recurrence | Y = 23 (39%) |
| N = 36 (61%) | |
| Death | Y = 54 (92%) |
| N = 5 (8%) | |
| mPFS (months) (95% CI; range) | 10.97 (6.51–15.43; 1.03–78.8) |
| mOS (months) (95% CI; range) | 18.87(15.3–22.44; 1.03 –82.2) |
| mFollow-up (months) (95% CI; range) | 78.7 (48.97–108.57; 1.03–82.2) |
| TILs Distribution | |||
|---|---|---|---|
| GBM | Normal-Appearing Tissue | p | |
| Mean CD4/HPF | 6.18 ± 6 (median 3.64) | 1.78 ± 2.7 (median 0.6) | <0.0001 |
| Mean CD8/HPF | 6.14 ± 8.91 (median 3.07) | 2.28 ± 2.17 (median 1.47) | 0.0005 |
| Mean CD4/CD8 | 1.86 ± 2.09 (median 1.23) | 1.26 ± 0.3 (median 0.31) | 0.0009 |
| Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|
| Overall Survival | Progression-Free Survival | |||||
| VARIABLE | p | HR | 95% CI | p | HR | 95% CI |
| Gender | 0.006 | 0.42 | 0.22–0.77 | 0.027 | 0.31 | 0.1–0.85 |
| Age | 0.019 | 2.15 | 1.15–4.12 | 0.048 | 0.37 | 0.13–0.97 |
| EOR | 0.129 | 1.58 | 0.88–2.88 | 0.141 | 0.46 | 0.15–1.24 |
| MGMT Met | 0.081 | 0.54 | 0.27–1.1 | 0.406 | 1.62 | 0.55–5.58 |
| RT | 0.767 | 0.77 | 0.42–1.84 | 0.004 | 0.07 | 0.01–0.35 |
| RT + CT | 0.769 | 0.77 | 0.42–1.9 | 0.038 | 5.61 | 1.28–39.04 |
| CD4L/CD8L | 0.014 | 0.38 | 0.18–0.79 | 0.495 | 1.43 | 0.44–5.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Innocenti, L.; Ortenzi, V.; Scarpitta, R.; Montemurro, N.; Pasqualetti, F.; Asseri, R.; Lazzi, S.; Szumera-Cieckiewicz, A.; De Ieso, K.; Perrini, P.; et al. The Prognostic Impact of Gender, Therapeutic Strategies, Molecular Background, and Tumor-Infiltrating Lymphocytes in Glioblastoma: A Still Unsolved Jigsaw. Genes 2023, 14, 501. https://doi.org/10.3390/genes14020501
Innocenti L, Ortenzi V, Scarpitta R, Montemurro N, Pasqualetti F, Asseri R, Lazzi S, Szumera-Cieckiewicz A, De Ieso K, Perrini P, et al. The Prognostic Impact of Gender, Therapeutic Strategies, Molecular Background, and Tumor-Infiltrating Lymphocytes in Glioblastoma: A Still Unsolved Jigsaw. Genes. 2023; 14(2):501. https://doi.org/10.3390/genes14020501
Chicago/Turabian StyleInnocenti, Lorenzo, Valerio Ortenzi, Rosa Scarpitta, Nicola Montemurro, Francesco Pasqualetti, Roberta Asseri, Stefano Lazzi, Anna Szumera-Cieckiewicz, Katia De Ieso, Paolo Perrini, and et al. 2023. "The Prognostic Impact of Gender, Therapeutic Strategies, Molecular Background, and Tumor-Infiltrating Lymphocytes in Glioblastoma: A Still Unsolved Jigsaw" Genes 14, no. 2: 501. https://doi.org/10.3390/genes14020501
APA StyleInnocenti, L., Ortenzi, V., Scarpitta, R., Montemurro, N., Pasqualetti, F., Asseri, R., Lazzi, S., Szumera-Cieckiewicz, A., De Ieso, K., Perrini, P., Naccarato, A. G., Scatena, C., & Fanelli, G. N. (2023). The Prognostic Impact of Gender, Therapeutic Strategies, Molecular Background, and Tumor-Infiltrating Lymphocytes in Glioblastoma: A Still Unsolved Jigsaw. Genes, 14(2), 501. https://doi.org/10.3390/genes14020501











