Phase Separation: Direct and Indirect Driving Force for High-Order Chromatin Organization
Abstract
1. Introduction
2. Snapshot of Liquid–Liquid Phase Separation
3. Current View of Spatial Chromatin Organization
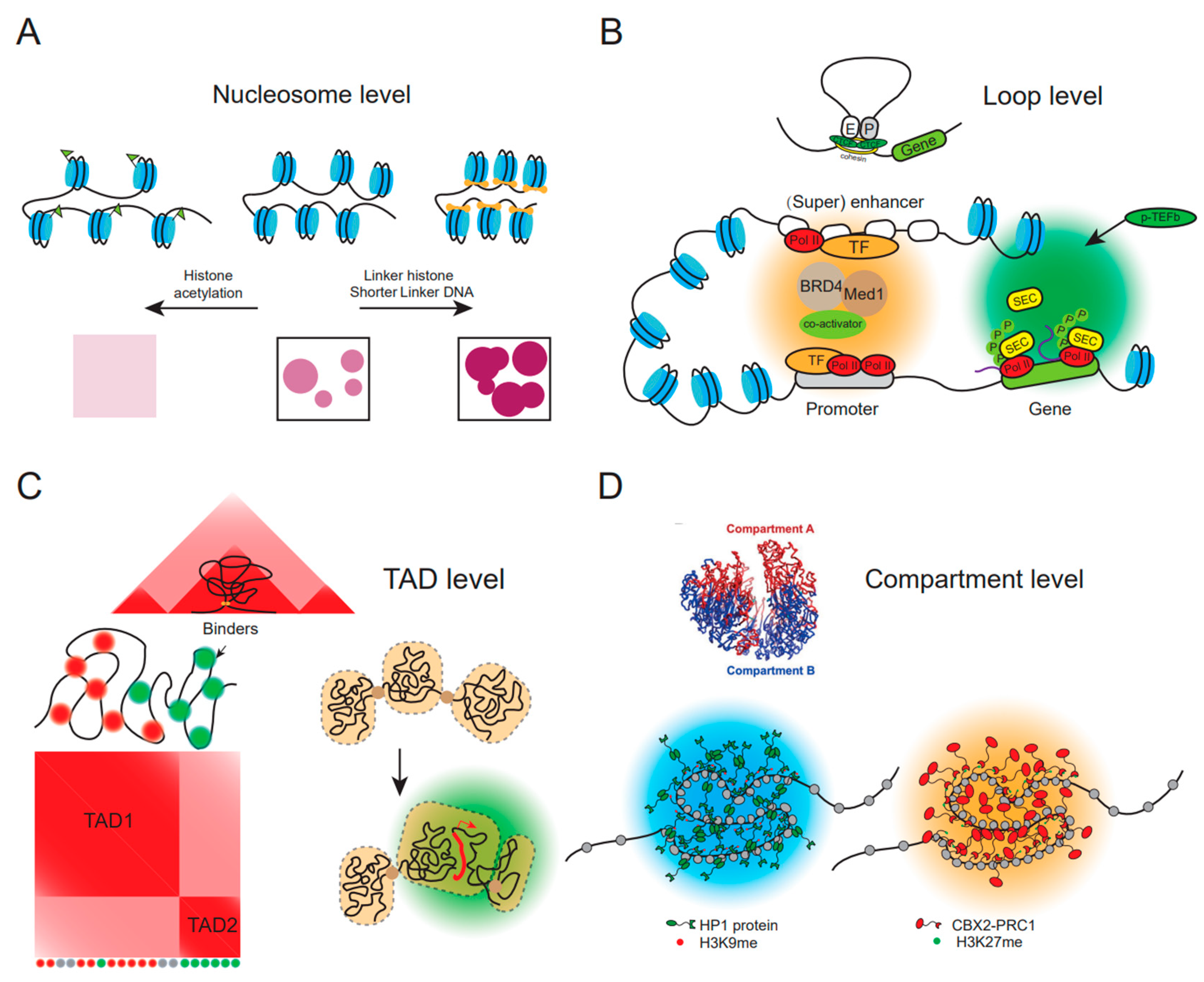
4. Phase Separation Directly Impacts All Layers of High-Order Chromatin Organization
4.1. Role of LLPS in Nucleosome-Mediated Chromatin Condensation
4.2. Role of LLPS in Chromatin Loop Formation
4.3. Role of Phase Separation in TAD Assembly and Reprogramming
4.4. Role of Phase Separation in Nuclear Compartmentalization
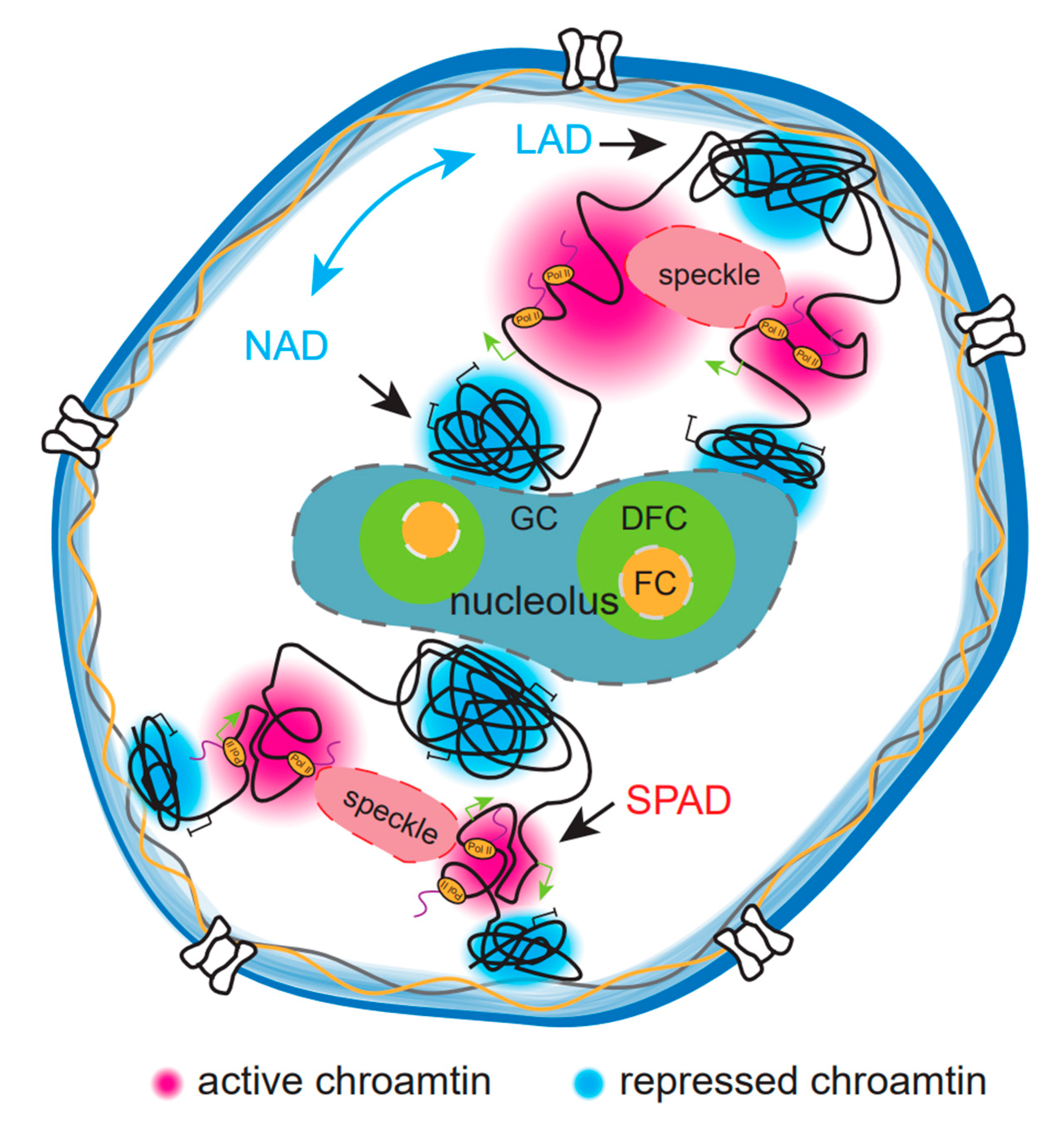
5. Indirect Role of Phase Separation Compartmentalization on Chromatin Organization
5.1. Nuclear Speckles and Chromatin Organization
5.2. Nucleolus and Chromatin Organization
5.3. Nuclear Lamina and Chromatin Organization
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Misteli, T. Beyond the sequence: Cellular organization of genome function. Cell 2007, 128, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Corces, V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Belmont, A.S. Nuclear Compartments: An Incomplete Primer to Nuclear Compartments, Bodies, and Genome Organization Relative to Nuclear Architecture. Cold Spring Harb. Perspect. Biol. 2022, 14, a041268. [Google Scholar] [CrossRef]
- Fawcett, D.W. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 1966, 119, 129–145. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef]
- Staněk, D.; Fox, A.H. Nuclear bodies: News insights into structure and function. Curr. Opin. Cell Biol. 2017, 46, 94–101. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Grosveld, F.G.; Papantonis, A. Forces driving the three-dimensional folding of eukaryotic genomes. Mol. Syst. Biol. 2018, 14, e8214. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Bhat, P.; Honson, D.; Guttman, M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Reviews. Mol. Cell Biol. 2021, 22, 653–670. [Google Scholar] [CrossRef]
- Li, J.; Gao, J.; Wang, R. Control of Chromatin Organization and Chromosome Behavior during the Cell Cycle through Phase Separation. Int. J. Mol. Sci. 2021, 22, 12271. [Google Scholar] [CrossRef] [PubMed]
- Pessina, F.; Gioia, U.; Brandi, O.; Farina, S.; Ceccon, M.; Francia, S.; d’Adda di Fagagna, F. DNA Damage Triggers a New Phase in Neurodegeneration. Trends Genet. TIG 2021, 37, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, E.M.; Dekker, J. Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem. Sci. 2020, 45, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef]
- Suzuki, H.; Abe, R.; Shimada, M.; Hirose, T.; Hirose, H.; Noguchi, K.; Ike, Y.; Yasui, N.; Furugori, K.; Yamaguchi, Y.; et al. The 3’ Pol II pausing at replication-dependent histone genes is regulated by Mediator through Cajal bodies’ association with histone locus bodies. Nat. Commun. 2022, 13, 2905. [Google Scholar] [CrossRef]
- Banjade, S.; Rosen, M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 2014, 3. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Nagulapalli, M.; Maji, S.; Dwivedi, N.; Dahiya, P.; Thakur, J.K. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 2016, 44, 1591–1612. [Google Scholar] [CrossRef]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Ströhl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173, 720–734.e715. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Chen, X.; Ma, L.; Li, P.; Yu, H. Protein phase separation and its role in chromatin organization and diseases. Biomed Pharm. 2021, 138, 111520. [Google Scholar] [CrossRef]
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Fullwood, M.J.; Wei, C.L.; Liu, E.T.; Ruan, Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009, 19, 521–532. [Google Scholar] [CrossRef]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef]
- Beagrie, R.A.; Scialdone, A.; Schueler, M.; Kraemer, D.C.; Chotalia, M.; Xie, S.Q.; Barbieri, M.; de Santiago, I.; Lavitas, L.M.; Branco, M.R.; et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 2017, 543, 519–524. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Beagan, J.A.; Phillips-Cremins, J.E. On the existence and functionality of topologically associating domains. Nat. Genet. 2020, 52, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, A.L.; Rao, S.S.; Huang, S.C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef] [PubMed]
- Vian, L.; Pękowska, A.; Rao, S.S.P.; Kieffer-Kwon, K.R.; Jung, S.; Baranello, L.; Huang, S.C.; El Khattabi, L.; Dose, M.; Pruett, N.; et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell 2018, 173, 1165–1178.e20. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Ulianov, S.V. Divide and Rule: Phase Separation in Eukaryotic Genome Functioning. Cells 2020, 9, 2480. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.L.; Watson, M.; Wilkins, O.G.; Cato, L.; Travers, A.; Thomas, J.O.; Stott, K. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl. Acad. Sci. USA 2018, 115, 11964–11969. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e21. [Google Scholar] [CrossRef]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 2017, 67, 837–852.e7. [Google Scholar] [CrossRef]
- Maeshima, K.; Rogge, R.; Tamura, S.; Joti, Y.; Hikima, T.; Szerlong, H.; Krause, C.; Herman, J.; Seidel, E.; DeLuca, J.; et al. Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 2016, 35, 1115–1132. [Google Scholar] [CrossRef]
- Gordon, F.; Luger, K.; Hansen, J.C. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 2005, 280, 33701–33706. [Google Scholar] [CrossRef] [PubMed]
- Shakya, A.; Park, S.; Rana, N.; King, J.T. Liquid-Liquid Phase Separation of Histone Proteins in Cells: Role in Chromatin Organization. Biophys. J. 2020, 118, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Ulianov, S.V.; Velichko, A.K.; Magnitov, M.D.; Luzhin, A.V.; Golov, A.K.; Ovsyannikova, N.; Kireev, I.; Gavrikov, A.S.; Mishin, A.S.; Garaev, A.K.; et al. Suppression of liquid-liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res. 2021, 49, 10524–10541. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Dong, L.; Tang, M.; Zhang, P.; Zhang, C.; Cao, Z.; Zhu, Q.; Chen, Y.; Wang, H.; et al. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018, 46, 7716–7730. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, L.; Quan, H.; Tian, H.; Meng, L.; Yang, L.; Feng, H.; Gao, Y.Q. From 1D sequence to 3D chromatin dynamics and cellular functions: A phase separation perspective. Nucleic Acids Res. 2018, 46, 9367–9383. [Google Scholar] [CrossRef]
- Fudenberg, G.; Abdennur, N.; Imakaev, M.; Goloborodko, A.; Mirny, L.A. Emerging Evidence of Chromosome Folding by Loop Extrusion. Cold Spring Harb. Symp. Quant. Biol. 2017, 82, 45–55. [Google Scholar] [CrossRef]
- Pugacheva, E.M.; Kubo, N.; Loukinov, D.; Tajmul, M.; Kang, S.; Kovalchuk, A.L.; Strunnikov, A.V.; Zentner, G.E.; Ren, B.; Lobanenkov, V.V. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 2020, 117, 2020–2031. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef]
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, eaar2555. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e16. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Che, Z.; Yue, J.; Xie, P.; Hao, S.; Xie, W.; Luo, Z.; Lin, C. ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Sci. Adv. 2020, 6, eaay4858. [Google Scholar] [CrossRef]
- Shin, Y.; Chang, Y.C.; Lee, D.S.W.; Berry, J.; Sanders, D.W.; Ronceray, P.; Wingreen, N.S.; Haataja, M.; Brangwynne, C.P. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 2018, 175, 1481–1491.e13. [Google Scholar] [CrossRef]
- Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H.; Li, J.; Storey, A.J.; Tsai, Y.H.; Keeley, D.P.; et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 2021, 595, 591–595. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, S.; Ma, L.; Qu, J.; Zhao, L.; Zhu, X.; Ding, J. Time-dependent effect of 1,6-hexanediol on biomolecular condensates and 3D chromatin organization. Genome Biol. 2021, 22, 230. [Google Scholar] [CrossRef]
- Crump, N.T.; Ballabio, E.; Godfrey, L.; Thorne, R.; Repapi, E.; Kerry, J.; Tapia, M.; Hua, P.; Lagerholm, C.; Filippakopoulos, P.; et al. BET inhibition disrupts transcription but retains enhancer-promoter contact. Nat. Commun. 2021, 12, 223. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719. [Google Scholar] [CrossRef]
- Zhang, Y.; See, Y.X.; Tergaonkar, V.; Fullwood, M.J. Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers. Cells 2022, 11, 1560. [Google Scholar] [CrossRef]
- Ngan, C.Y.; Wong, C.H.; Tjong, H.; Wang, W.; Goldfeder, R.L.; Choi, C.; He, H.; Gong, L.; Lin, J.; Urban, B.; et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat. Genet. 2020, 52, 264–272. [Google Scholar] [CrossRef]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Finn, E.H.; Pegoraro, G.; Brandao, H.B.; Valton, A.L.; Oomen, M.E.; Dekker, J.; Mirny, L.; Misteli, T. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 2019, 176, 1502–1515.e1510. [Google Scholar] [CrossRef] [PubMed]
- Bintu, B.; Mateo, L.J.; Su, J.H.; Sinnott-Armstrong, N.A.; Parker, M.; Kinrot, S.; Yamaya, K.; Boettiger, A.N.; Zhuang, X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 2018, 362, eaau1783. [Google Scholar] [CrossRef] [PubMed]
- Esposito, A.; Bianco, S.; Fiorillo, L.; Conte, M.; Abraham, A.; Musella, F.; Nicodemi, M.; Prisco, A.; Chiariello, A.M. Polymer models are a versatile tool to study chromatin 3D organization. Biochem. Soc. Trans. 2021, 49, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, M.; Chotalia, M.; Fraser, J.; Lavitas, L.M.; Dostie, J.; Pombo, A.; Nicodemi, M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA 2012, 109, 16173–16178. [Google Scholar] [CrossRef]
- Miron, E.; Oldenkamp, R.; Brown, J.M.; Pinto, D.M.S.; Xu, C.S.; Faria, A.R.; Shaban, H.A.; Rhodes, J.D.P.; Innocent, C.; de Ornellas, S.; et al. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv. 2020, 6, eaba8811. [Google Scholar] [CrossRef]
- Conte, M.; Fiorillo, L.; Bianco, S.; Chiariello, A.M.; Esposito, A. and Nicodemi, M. Polymer Physics Indicates Chro-matin Folding Variability across Single-Cells Results from State Degeneracy in Phase Separation. Nat. Commun. 2020, 11, 3289. [Google Scholar] [CrossRef]
- Conte, M.; Irani, E.; Chiariello, A.M.; Abraham, A.; Bianco, S.; Esposito, A.; Nicodemi, M. Loop-extrusion and polymer phase-separation can co-exist at the single-molecule level to shape chromatin folding. Nat. Commun. 2022, 13, 4070. [Google Scholar] [CrossRef]
- Ryu, J.K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; de Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci. Adv. 2021, 7, eabe5905. [Google Scholar] [CrossRef]
- Esposito, A.; Abraham, A.; Conte, M.; Vercellone, F.; Prisco, A.; Bianco, S.; Chiariello, A.M. The Physics of DNA Folding: Polymer Models and Phase-Separation. Polymers 2022, 14, 1918. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Ma, Q.; Zeng, P.; Wu, D.; Hou, Y.; Liu, X.; Jia, L.; Sun, J.; Chen, Y.; et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 2021, 28, 1868–1883.e1811. [Google Scholar] [CrossRef]
- Erdel, F. Biophysical mechanisms of chromatin patterning. Curr. Opin. Genet. Dev. 2020, 61, 62–68. [Google Scholar] [CrossRef]
- McSwiggen, D.T.; Hansen, A.S.; Teves, S.S.; Marie-Nelly, H.; Hao, Y.; Heckert, A.B.; Umemoto, K.K.; Dugast-Darzacq, C.; Tjian, R.; Darzacq, X. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 2019, 8, e47098. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Y.; Zheng, X.; Liu, C.; Dong, S.; Li, R.; Zhang, G.; Wei, Y.; Qu, H.; Li, Y.; et al. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol. Cell 2019, 76, 646–659.e6. [Google Scholar] [CrossRef]
- Olvera de la Cruz, M. Theory of microphase separation in block copolymer solutions. J. Chem. Phys. 1989, 90, 1995–2002. [Google Scholar] [CrossRef]
- Ryu, J.K.; Hwang, D.E.; Choi, J.M. Current Understanding of Molecular Phase Separation in Chromosomes. Int. J. Mol. Sci. 2021, 22, 10736. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Sanulli, S.; Trnka, M.J.; Dharmarajan, V.; Tibble, R.W.; Pascal, B.D.; Burlingame, A.L.; Griffin, P.R.; Gross, J.D.; Narlikar, G.J. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019, 575, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Oki, M.; Masai, H. Regulation of HP1 protein by phosphorylation during transcriptional repression and cell cycle. J. Biochem. 2021, 169, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Zenk, F.; Zhan, Y.; Kos, P.; Loser, E.; Atinbayeva, N.; Schachtle, M.; Tiana, G.; Giorgetti, L.; Iovino, N. HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature 2021, 593, 289–293. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef]
- Plys, A.J.; Davis, C.P.; Kim, J.; Rizki, G.; Keenen, M.M.; Marr, S.K.; Kingston, R.E. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Kent, S.; Brown, K.; Yang, C.H.; Alsaihati, N.; Tian, C.; Wang, H.; Ren, X. Phase-Separated Transcriptional Condensates Accelerate Target-Search Process Revealed by Live-Cell Single-Molecule Imaging. Cell Rep. 2020, 33, 108248. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yu, D.; Gu, R.; Jia, Y.; Wang, Q.; Jaganathan, A.; Yang, X.; Yu, M.; Babault, N.; Zhao, C.; et al. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat. Struct. Mol. Biol. 2020, 27, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Eeftens, J.M.; Kapoor, M.; Michieletto, D.; Brangwynne, C.P. Polycomb condensates can promote epigenetic marks but are not required for sustained chromatin compaction. Nat. Commun. 2021, 12, 5888. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Jia, L.; Huang, X.; Tan, J.; Wang, M.; Niu, J.; Hou, Y.; Sun, J.; Zeng, P.; Wang, J.; et al. CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation. Cell Res. 2022, 32, 744–760. [Google Scholar] [CrossRef]
- Pliss, A.; Peng, X.; Liu, L.; Kuzmin, A.; Wang, Y.; Qu, J.; Li, Y.; Prasad, P.N. Single Cell Assay for Molecular Diagnostics and Medicine: Monitoring Intracellular Concentrations of Macromolecules by Two-photon Fluorescence Lifetime Imaging. Theranostics 2015, 5, 919–930. [Google Scholar] [CrossRef]
- Ilik, İ.A.; Malszycki, M.; Lübke, A.K.; Schade, C.; Meierhofer, D.; Aktaş, T. SON and SRRM2 are essential for nuclear speckle formation. eLife 2020, 9, e60579. [Google Scholar] [CrossRef]
- Hall, L.L.; Smith, K.P.; Byron, M.; Lawrence, J.B. Molecular anatomy of a speckle. Anat. Record. Part A Discov. Mol. Cell. Evol. Biol. 2006, 288, 664–675. [Google Scholar] [CrossRef]
- Misteli, T.; Cáceres, J.F.; Spector, D.L. The dynamics of a pre-mRNA splicing factor in living cells. Nature 1997, 387, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018, 217, 4025–4048. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yan, Z.; Li, S.; Huang, N.; Huang, X.; Zhang, J.; Zhong, S. RNAs as Proximity-Labeling Media for Identifying Nuclear Speckle Positions Relative to the Genome. iScience 2018, 4, 204–215. [Google Scholar] [CrossRef]
- Ahanger, S.H.; Delgado, R.N.; Gil, E.; Cole, M.A.; Zhao, J.; Hong, S.J.; Kriegstein, A.R.; Nowakowski, T.J.; Pollen, A.A.; Lim, D.A. Distinct nuclear compartment-associated genome architecture in the developing mammalian brain. Nat. Neurosci. 2021, 24, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Dow, E.C.; Liu, H.; Rice, A.P. T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes. J. Cell. Physiol. 2010, 224, 84–93. [Google Scholar] [CrossRef]
- Hu, S.; Lv, P.; Yan, Z.; Wen, B. Disruption of nuclear speckles reduces chromatin interactions in active compartments. Epigenetics Chromatin 2019, 12, 43. [Google Scholar] [CrossRef]
- Khanna, N.; Hu, Y.; Belmont, A.S. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. CB 2014, 24, 1138–1144. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J. Birth of Nucleolar Compartments: Phase Separation-Driven Ribosomal RNA Sorting and Processing. Mol. Cell 2019, 76, 694–696. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Girard, J.P.; Lehtonen, H.; Caizergues-Ferrer, M.; Amalric, F.; Tollervey, D.; Lapeyre, B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992, 11, 673–682. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Grace, C.R.; Buljan, M.; Yun, M.K.; Pytel, N.J.; Satumba, J.; Nourse, A.; Park, C.G.; Madan Babu, M.; White, S.W.; et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl. Acad. Sci. USA 2014, 111, 4466–4471. [Google Scholar] [CrossRef] [PubMed]
- Colau, G.; Thiry, M.; Leduc, V.; Bordonné, R.; Lafontaine, D.L. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol. Cell. Biol. 2004, 24, 7976–7986. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Weber, S.C.; Vaidya, N.; Haataja, M.; Brangwynne, C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. USA 2015, 112, E5237–E5245. [Google Scholar] [CrossRef]
- Mangan, H.; Gailín, M.; McStay, B. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017, 284, 3977–3985. [Google Scholar] [CrossRef] [PubMed]
- Weierich, C.; Brero, A.; Stein, S.; von Hase, J.; Cremer, C.; Cremer, T.; Solovei, I. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2003, 11, 485–502. [Google Scholar] [CrossRef]
- Zhang, S.; Hemmerich, P.; Grosse, F. Nucleolar localization of the human telomeric repeat binding factor 2 (TRF2). J. Cell Sci. 2004, 117, 3935–3945. [Google Scholar] [CrossRef]
- Zhang, L.F.; Huynh, K.D.; Lee, J.T. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell 2007, 129, 693–706. [Google Scholar] [CrossRef]
- van Koningsbruggen, S.; Gierlinski, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y.; den Dunnen, J.T.; Lamond, A.I. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 2010, 21, 3735–3748. [Google Scholar] [CrossRef]
- Bersaglieri, C.; Kresoja-Rakic, J.; Gupta, S.; Bar, D.; Kuzyakiv, R.; Panatta, M.; Santoro, R. Genome-wide maps of nucleolus interactions reveal distinct layers of repressive chromatin domains. Nat. Commun. 2022, 13, 1483. [Google Scholar] [CrossRef]
- Padeken, J.; Mendiburo, M.J.; Chlamydas, S.; Schwarz, H.J.; Kremmer, E.; Heun, P. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol. Cell 2013, 50, 236–249. [Google Scholar] [CrossRef]
- Vertii, A.; Ou, J.; Yu, J.; Yan, A.; Pagès, H.; Liu, H.; Zhu, L.J.; Kaufman, P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019, 29, 1235–1249. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Serebryannyy, L.; Misteli, T. Protein sequestration at the nuclear periphery as a potential regulatory mechanism in premature aging. J. Cell Biol. 2018, 217, 21–37. [Google Scholar] [CrossRef] [PubMed]
- van Steensel, B.; Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef]
- Gupta, S.; Santoro, R. Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization. Stem Cell Rep. 2020, 15, 1206–1219. [Google Scholar] [CrossRef]
- Nazer, E. To be or not be (in the LAD): Emerging roles of lamin proteins in transcriptional regulation. Biochem. Soc. Trans. 2022, 50, 1035–1044. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, J.; Yue, S.; Kristiani, L.; Kim, M.; Sauria, M.; Taylor, J.; Kim, Y.; Zheng, Y. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 2018, 71, 802–815.e7. [Google Scholar] [CrossRef]
- Falk, M.; Feodorova, Y.; Naumova, N.; Imakaev, M.; Lajoie, B.R.; Leonhardt, H.; Joffe, B.; Dekker, J.; Fudenberg, G.; Solovei, I.; et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019, 570, 395–399. [Google Scholar] [CrossRef]
- Karoutas, A.; Akhtar, A. Functional mechanisms and abnormalities of the nuclear lamina. Nat. Cell Biol. 2021, 23, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Callebaut, I.; Pezhman, A.; Courvalin, J.C.; Worman, H.J. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997, 272, 14983–14989. [Google Scholar] [CrossRef] [PubMed]
- See, K.; Kiseleva, A.A.; Smith, C.L.; Liu, F.; Li, J.; Poleshko, A.; Epstein, J.A. Histone methyltransferase activity programs nuclear peripheral genome positioning. Dev. Biol. 2020, 466, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sandoval, A.; Towbin, B.D.; Kalck, V.; Cabianca, D.S.; Gaidatzis, D.; Hauer, M.H.; Geng, L.; Wang, L.; Yang, T.; Wang, X.; et al. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015, 163, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.G.; Han, Z.Z.; Daude, N.; McNamara, E.; Wohlgemuth, S.; Molina-Porcel, L.; Safar, J.G.; Mok, S.A.; Westaway, D. Pathologic tau conformer ensembles induce dynamic, liquid-liquid phase separation events at the nuclear envelope. BMC Biol. 2021, 19, 199. [Google Scholar] [CrossRef]
- Dormann, D. FG-nucleoporins caught in the act of liquid-liquid phase separation. J. Cell Biol. 2020, 219, e201910211. [Google Scholar] [CrossRef]
- Wang, Q.; Sawyer, I.A.; Sung, M.H.; Sturgill, D.; Shevtsov, S.P.; Pegoraro, G.; Hakim, O.; Baek, S.; Hager, G.L.; Dundr, M. Cajal bodies are linked to genome conformation. Nat. Commun. 2016, 7, 10966. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef]
- Shi, M.; You, K.; Chen, T.; Hou, C.; Liang, Z.; Liu, M.; Wang, J.; Wei, T.; Qin, J.; Chen, Y.; et al. Quantifying the phase separation property of chromatin-associated proteins under physiological conditions using an anti-1,6-hexanediol index. Genome Biol. 2021, 22, 229. [Google Scholar] [CrossRef]
- Ling, X.; Liu, X.; Jiang, S.; Fan, L.; Ding, J. The dynamics of three-dimensional chromatin organization and phase separation in cell fate transitions and diseases. Cell Regen. 2022, 11, 42. [Google Scholar] [CrossRef]
- Wang, L.; Hu, M.; Zuo, M.Q.; Zhao, J.; Wu, D.; Huang, L.; Wen, Y.; Li, Y.; Chen, P.; Bao, X.; et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 2020, 30, 393–407. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; An, Z.; Zhang, W.; Li, F. Phase Separation: Direct and Indirect Driving Force for High-Order Chromatin Organization. Genes 2023, 14, 499. https://doi.org/10.3390/genes14020499
Li X, An Z, Zhang W, Li F. Phase Separation: Direct and Indirect Driving Force for High-Order Chromatin Organization. Genes. 2023; 14(2):499. https://doi.org/10.3390/genes14020499
Chicago/Turabian StyleLi, Xiaoli, Ziyang An, Wenqing Zhang, and Feifei Li. 2023. "Phase Separation: Direct and Indirect Driving Force for High-Order Chromatin Organization" Genes 14, no. 2: 499. https://doi.org/10.3390/genes14020499
APA StyleLi, X., An, Z., Zhang, W., & Li, F. (2023). Phase Separation: Direct and Indirect Driving Force for High-Order Chromatin Organization. Genes, 14(2), 499. https://doi.org/10.3390/genes14020499




