Informing Wildlife Corridor Creation through Population Genetics of an Arboreal Marsupial in a Fragmented Landscape
Abstract
1. Introduction
2. Methods Section
2.1. Study Area
2.2. Live Trapping and Tissue Collection
2.3. SNP Genotyping and Filtering
2.4. Genetic Diversity Analyses
2.5. Population Genetic Structure Analyses
2.6. Isolation by Distance Analyses
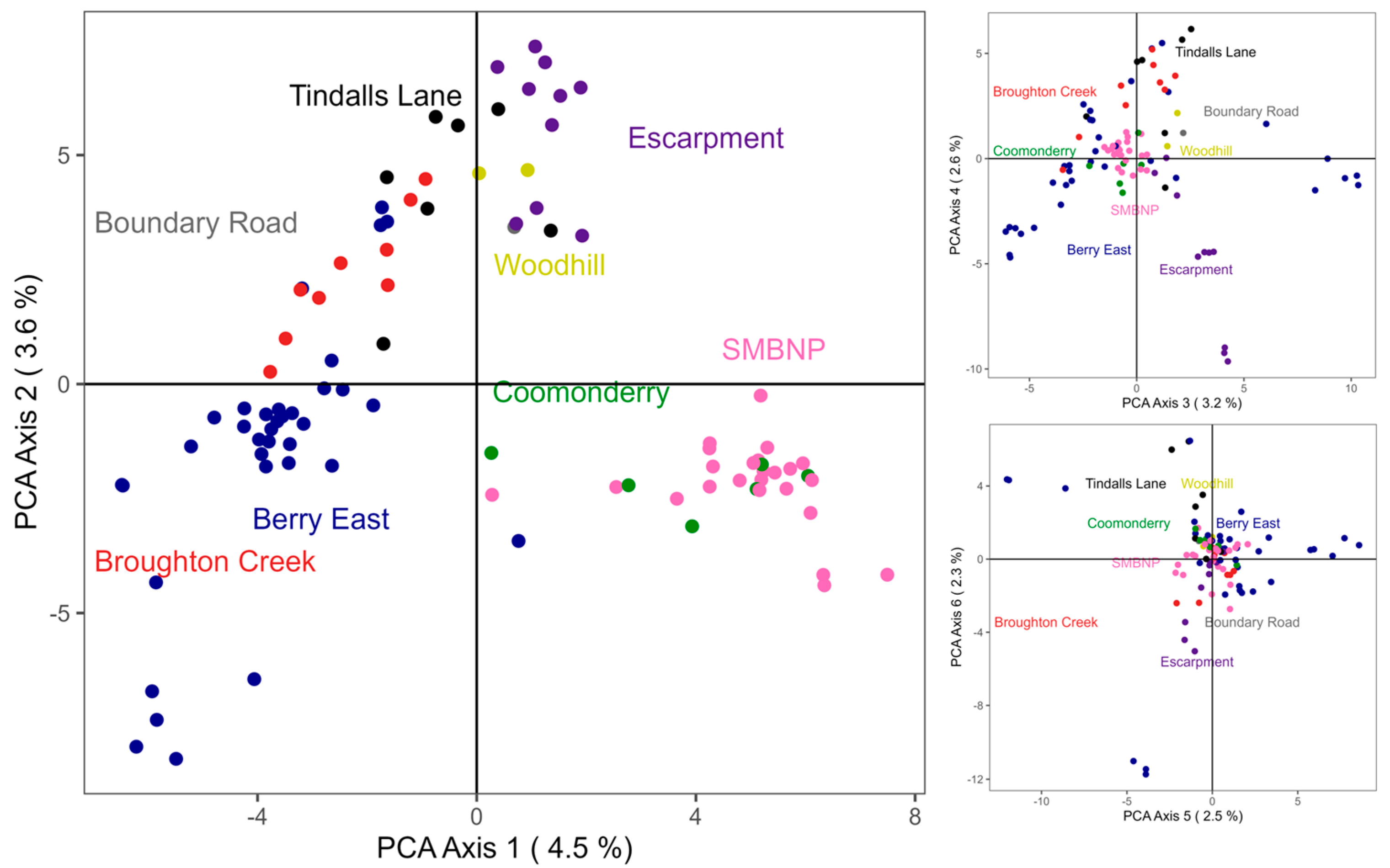
3. Results
3.1. Genetic Diversity
3.2. Population Structure
3.3. Isolation by Distance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Powers, R.P.; Jetz, W. Global Habitat Loss and Extinction Risk of Terrestrial Vertebrates under Future Land-Use-Change Scenarios. Nat. Clim. Chang. 2019, 9, 323–329. [Google Scholar] [CrossRef]
- Schipper, J.; Chanson, J.S.; Chiozza, F.; Cox, N.A.; Hoffmann, M.; Katariya, V.; Lamoreux, J.; Rodrigues, A.S.L.; Stuart, S.N.; Temple, H.J.; et al. The Status of the World’s Land and Marine Mammals: Diversity, Threat, and Knowledge. Science 2008, 322, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat Fragmentation and Its Lasting Impact on Earth’s Ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Chen, X.Y.; Corlett, R.T.; Didham, R.K.; Ding, P.; Holt, R.D.; Holyoak, M.; Hu, G.; Hughes, A.C.; Jiang, L.; et al. Habitat Fragmentation and Biodiversity Conservation: Key Findings and Future Challenges. Landsc. Ecol. 2016, 31, 219–227. [Google Scholar] [CrossRef]
- Gilpin, M.E.; Soule, M.E. Minimum Viable Populations: Processes of Species Extinction. In Conservation Biology: The Science of Scarcity and Diversity; Oxford University Press: New York, NY, USA, 1986; ISBN 9780791850503. [Google Scholar]
- Fagan, W.F.; Holmes, E. Quantifying the Extinction Vortex. Ecol. Lett. 2006, 9, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Brook, B.W.; Sodhi, N.S.; Bradshaw, C.J.A. Synergies among Extinction Drivers under Global Change. Trends Ecol. Evol. 2008, 23, 453–460. [Google Scholar] [CrossRef]
- Keinath, D.A.; Doak, D.F.; Hodges, K.E.; Prugh, L.R.; Fagan, W.; Sekercioglu, C.H.; Buchart, S.H.M.; Kauffman, M. A Global Analysis of Traits Predicting Species Sensitivity to Habitat Fragmentation. Glob. Ecol. Biogeogr. 2017, 26, 115–127. [Google Scholar] [CrossRef]
- Koprowski, J.L. The Response of Tree Squirrels to Fragmentation: A Review and Synthesis. Anim. Conserv. 2005, 8, 369–376. [Google Scholar] [CrossRef]
- Isaac, B.; White, J.; Ierodiaconou, D.; Cooke, R. Simplification of Arboreal Marsupial Assemblages in Response to Increasing Urbanization. PLoS ONE 2014, 9, e91049. [Google Scholar] [CrossRef]
- Fietz, J.; Weis-Dootz, T. Stranded on an Island: Consequences of Forest Fragmentation for Body Size Variations in an Arboreal Mammal, the Edible Dormouse (Glis Glis). Popul. Ecol. 2012, 54, 313–320. [Google Scholar] [CrossRef]
- Wauters, L.A.; Dhondt, A.A.; Knothe, H.; Parkin, D.T. Fluctuating Asymmetry and Body Size as Indicators of Stress in Red Squirrel Populations in Woodland Fragments. J. Appl. Ecol. 1996, 33, 735–740. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010; ISBN 0521702712. [Google Scholar]
- Lancaster, M.L.; Cooper, S.J.B.; Carthew, S.M. Genetic Consequences of Forest Fragmentation by Agricultural Land in an Arboreal Marsupial. Landsc. Ecol. 2016, 31, 655–667. [Google Scholar] [CrossRef]
- Ramos Pereira, M.J.; Rojas, D.; Lino, A.; Fonseca, C.; Fischer, E. A Meta-Analysis of the Effects of Habitat Loss and Fragmentation on Genetic Diversity in Mammals. Mamm. Biol. 2018, 94, 69–76. [Google Scholar] [CrossRef]
- Knipler, M.; Dowton, M.; Clulow, J.; Meyer, N.; Mikac, K.M. Genome-Wide SNPs Detect Fine-Scale Genetic Structure in Threatened Populations of Squirrel Glider Petaurus norfolcensis. Conserv. Genet. 2022, 23, 541–558. [Google Scholar] [CrossRef]
- Lacy, R.C. Impacts of Inbreeding in Natural and Captive Populations of Vertebrates: Implications for Conservation. Perspect. Biol. Med. 1993, 36, 480–496. [Google Scholar] [CrossRef]
- Weeks, A.R.; Stoklosa, J.; Hoffmann, A.A. Conservation of Genetic Uniqueness of Populations May Increase Extinction Likelihood of Endangered Species: The Case of Australian Mammals. Front. Zool. 2016, 13, 31. [Google Scholar] [CrossRef]
- Willi, Y.; van Buskirk, J.; Schmid, B.; Fischer, M. Genetic Isolation of Fragmented Populations Is Exacerbated by Drift and Selection. J. Evol. Biol. 2007, 20, 534–542. [Google Scholar] [CrossRef]
- Reed, D.H. Extinction Risk in Fragmented Habitats. Anim. Conserv. 2004, 7, 181–191. [Google Scholar] [CrossRef]
- Beier, P.; Noss, R.F. Do Habitat Corridors Provide Connectivity? Conserv. Biol. 1998, 12, 1241–1252. [Google Scholar] [CrossRef]
- Hilty, J.A.; Lidicker, W.Z., Jr.; Merenlender, A.M. Corridor Ecology: The Science and Practice of Linking Landscapes for Biodiversity Conservation; Island Press: Washington, DC, USA, 2012; ISBN 1597265934. [Google Scholar]
- Bennett, A. Linkages in the Landscape. In The Role of Corridors and Connectivity in Wildlife Conservation, 2nd ed.; IUCN, The World Conservation Union: Gland, Switzerland, 2003. [Google Scholar]
- Wilson, R.F.; Marsh, H.; Winter, J. Importance of Canopy Connectivity for Home Range and Movements of the Rainforest Arboreal Ringtail Possum (Hemibelideus lemuroides). Wildl. Res. 2007, 34, 177–184. [Google Scholar] [CrossRef]
- Soanes, K.; Taylor, A.C.; Sunnucks, P.; Esk, P.A.; Cesarini, S.; van der Ree, R. Evaluating the Success of Wildlife Crossing Structures Using Genetic Approaches and an Experimental Design: Lessons from a Gliding Mammal. J. Appl. Ecol. 2017, 55, 129–138. [Google Scholar] [CrossRef]
- Jackson, S.M.; Parsons, M.; Baseler, M.; Stanton, D. Landscape Management of the Mahogany Glider (Petaurus gracilis) across Its Distribution: Subpopulations and Corridor Priorities. Aust. Mammal. 2020, 42, 152–159. [Google Scholar] [CrossRef]
- Mech, S.G.; Hallett, J.G. Evaluating the Effectiveness of Corridors: A Genetic Approach. Conserv. Biol. 2001, 15, 467–474. [Google Scholar] [CrossRef]
- Burkart, S.; Gugerli, F.; Senn, J.; Kuehn, R.; Bolliger, J. Evaluating the Functionality of Expert-Assessed Wildlife Corridors with Genetic Data from Roe Deer. Basic Appl. Ecol. 2016, 17, 52–60. [Google Scholar] [CrossRef]
- Keyghobadi, N. The Genetic Implications of Habitat Fragmentation for Animals. Can J Zool 2007, 85, 1049–1064. [Google Scholar] [CrossRef]
- Moraes, A.M.; Ruiz-Miranda, C.R.; Galetti, P.M., Jr.; Niebuhr, B.B.; Alexandre, B.R.; Muylaert, R.L.; Grativol, A.D.; Ribeiro, J.W.; Ferreira, A.N.; Ribeiro, M.C. Landscape Resistance Influences Effective Dispersal of Endangered Golden Lion Tamarins within the Atlantic Forest. Biol. Conserv. 2018, 224, 178–187. [Google Scholar] [CrossRef]
- Fraser, D.L.; Ironside, K.; Wayne, R.K.; Boydston, E.E. Connectivity of Mule Deer (Odocoileus hemionus) Populations in a Highly Fragmented Urban Landscape. Landsc. Ecol. 2019, 34, 1097–1115. [Google Scholar] [CrossRef]
- Fenderson, L.E.; Kovach, A.I.; Llamas, B. Spatiotemporal Landscape Genetics: Investigating Ecology and Evolution through Space and Time. Mol. Ecol. 2020, 29, 218–246. [Google Scholar] [CrossRef]
- Barbosa, S.; Mestre, F.; White, T.A.; Paupério, J.; Alves, P.C.; Searle, J.B. Integrative Approaches to Guide Conservation Decisions: Using Genomics to Define Conservation Units and Functional Corridors. Mol. Ecol. 2018, 27, 3452–3465. [Google Scholar] [CrossRef]
- Balkenhol, N.; Waits, L.P. Molecular Road Ecology: Exploring the Potential of Genetics for Investigating Transportation Impacts on Wildlife. Mol. Ecol. 2009, 18, 4151–4164. [Google Scholar] [CrossRef]
- Conner, M.M.; Saunders, W.C.; Bouwes, N.; Jordan, C. Evaluating Impacts Using a BACI Design, Ratios, and a Bayesian Approach with a Focus on Restoration. Environ. Monit. Assess. 2016, 188, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Suckling, G.C. Population Ecology of the Sugar Glider, Petaurus breviceps, in a System of Fragmented Habitats. Wildl. Res. 1984, 11, 49–75. [Google Scholar] [CrossRef]
- Caryl, F.M.; Thomson, K.; van der Ree, R. Permeability of the Urban Matrix to Arboreal Gliding Mammals: Sugar Gliders in Melbourne, Australia. Austral Ecol. 2013, 38, 609–613. [Google Scholar] [CrossRef]
- Jackson, S.M. Glide Angle in the Genus Petaurus and a Review of Gliding in Mammals. Mammal Rev. 2000, 30, 9–30. [Google Scholar] [CrossRef]
- Knipler, M.; Dowton, M.; Mikac, K. Limited Genetic Structure Detected in Sugar Gliders (Petaurus breviceps) Using Genome-Wide SNPs. Aust. Mammal. 2022, 45, 41–52. [Google Scholar] [CrossRef]
- Malekian, M.; Cooper, S.J.B.B.; Saint, K.M.; Lancaster, M.L.; Taylor, A.C.; Carthew, S.M.; Lancaster, M.L.; Malekian, M.; Carthew, S.M.; Cooper, S.J.B.B.; et al. Effects of Landscape Matrix on Population Connectivity of an Arboreal Mammal, Petaurus breviceps. Ecol. Evol. 2015, 5, 3939–3953. [Google Scholar] [CrossRef]
- Taylor, B.D.; Rohweder, D. Radio-Tracking Three Sugar Gliders Using Forested Highway Median Strips at Bongil Bongil National Park, North-East New South Wales. Ecol. Manag. Restor. 2013, 14, 228–230. [Google Scholar] [CrossRef]
- Shannon, M. Photopollution Impacts on the Nocturnal Behaviour of the Sugar Glider (Petaurus breviceps). Pac. Conserv. Biol. 2007, 13, 171–176. [Google Scholar]
- Smith, A.P. Diet and Feeding Strategies of the Marsupial Sugar Glider in Temperate Australia. J. Anim. Ecol. 1982, 51, 149. [Google Scholar] [CrossRef]
- Stojanovic, D.; Webb, M.H.; Alderman, R.; Porfirio, L.L.; Heinsohn, R. Discovery of a Novel Predator Reveals Extreme but Highly Variable Mortality for an Endangered Migratory Bird. Divers. Distrib. 2014, 20, 1200–1207. [Google Scholar] [CrossRef]
- Gracanin, A.; Cappelletti, C.; Knipler, M.; Dallas, R.K.K.; Mikac, K.M. Exploring New Grounds: Arboreal Sugar Gliders Frequently Observed Spending Time on the Ground as Seen on Camera Traps. Aust. Mammal. 2019, 42, 10–13. [Google Scholar] [CrossRef]
- GER The Great Eastern Ranges. Available online: https://ger.org.au/ (accessed on 23 March 2021).
- Perry, T.M. Biography—Alexander Berry—Australian Dictionary of Biography. Available online: https://adb.anu.edu.au/biography/berry-alexander-1773/text1987 (accessed on 5 December 2022).
- Gracanin, A.; Mikac, K.M. The Use of Selfie Camera Traps to Estimate Home Range and Movement Patterns of Small Mammals in a Fragmented Landscape. Animals 2022, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Tasker, E.M.; Dickman, C.R. A Review of Elliott Trapping Methods for Small Mammals in Australia. Aust. Mammal. 2001, 23, 77–87. [Google Scholar] [CrossRef]
- Suckling, G.C.; Macfarlane, M.A. Introduction of the Sugar Glider, Petaurus breviceps, into Re-Established Forest of the Tower Hill State Game Reserve, Vic. Wildl. Res. 1983, 10, 249–258. [Google Scholar] [CrossRef]
- Jackson, S.M. Home-Range and Den Use of the Mahogany Glider, Petaurus gracilis. Wildl. Res. 2000, 27, 49–60. [Google Scholar] [CrossRef]
- Sharpe, D.J.; Goldingay, R.L. Home Range of the Australian Squirrel Glider, Petaurus norfolcensis (Diprotodontia). J. Mammal. 2007, 88, 1515–1522. [Google Scholar] [CrossRef]
- Nowack, J.; Rojas, A.D.; Körtner, G.; Geiser, F. Snoozing through the Storm: Torpor Use during a Natural Disaster. Sci. Rep. 2015, 5, 11243. [Google Scholar] [CrossRef]
- Knipler, M.L.; Dowton, M.; Mikac, K.M. Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps Breviceps × Petaurus norfolcensis) in the Wild. Genes 2021, 12, 1327. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity Arrays Technology: A Generic Genome Profiling Technology on Open Platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-Project.org/ (accessed on 15 February 2019).
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. Dartr: An r Package to Facilitate Analysis of SNP Data Generated from Reduced Representation Genome Sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef]
- Privé, F.; Luu, K.; Vilhjálmsson, B.J.; Blum, M.G.B. Performing Highly Efficient Genome Scans for Local Adaptation with R Package Pcadapt Version 4. Mol. Biol. Evol. 2020, 37, 2153–2154. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. HIERFSTAT, a Package for R to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Dray, S.; Siberchicot, M.A. Package ‘Ade4’; Université de Lyon: Lyon, France, 2017. [Google Scholar]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant Analysis of Principal Components: A New Method for the Analysis of Genetically Structured Populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.F.; Franco, F.F.; Bombonato, J.R.; Bonatelli, I.A.S.; Khan, G.; Romeiro-Brito, M.; Fegies, A.C.; Ribeiro, P.M.; Silva, G.A.R.; Moraes, E.M. Assessing Population Structure in the Face of Isolation by Distance: Are We Neglecting the Problem? Divers. Distrib. 2018, 24, 1883–1889. [Google Scholar] [CrossRef]
- Wright, S. Isolation by Distance. Genetics 1943, 28, 114. [Google Scholar] [CrossRef]
- Gracanin, A.; Mikac, K.M. Evaluating Modelled Wildlife Corridors for the Movement of Multiple Arboreal Species in a Fragmented Landscape. Landsc. Ecol. 2022. submitted. [Google Scholar]
- Wang, Y.-H.; Yang, K.-C.; Bridgman, C.L.; Lin, L.-K. Habitat Suitability Modelling to Correlate Gene Flow with Landscape Connectivity. Landsc. Ecol. 2008, 23, 989–1000. [Google Scholar] [CrossRef]
- Lancaster, M.L.; Taylor, A.C.; Cooper, S.J.B.; Carthew, S.M. Limited Ecological Connectivity of an Arboreal Marsupial across a Forest/Plantation Landscape despite Apparent Resilience to Fragmentation. Mol. Ecol. 2011, 20, 2258–2271. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.C.; Walker, F.M.; Goldingay, R.L.; Ball, T.; van der Ree, R. Degree of Landscape Fragmentation Influences Genetic Isolation among Populations of a Gliding Mammal. PLoS ONE 2011, 6, e26651. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.D.; Sharpe, D.J.; Goldingay, R.L.; Ball, T.M.; Taylor, A.C.; Harrisson, K.A. Fine-Scale Genetic Response to Landscape Change in a Gliding Mammal. PLoS ONE 2013, 8, e80383. [Google Scholar] [CrossRef]
- Yokochi, K.; Kennington, W.J.; Bencini, R. An Endangered Arboreal Specialist, the Western Ringtail Possum (Pseudocheirus occidentalis), Shows a Greater Genetic Divergence across a Narrow Artificial Waterway than a Major Road. PLoS ONE 2016, 11, e0146167. [Google Scholar] [CrossRef] [PubMed]
- Fietz, J.; Tomiuk, J.; Loeschcke, V.; Weis-Dootz, T.; Segelbacher, G. Genetic Consequences of Forest Fragmentation for a Highly Specialized Arboreal Mammal—The Edible Dormouse. PLoS ONE 2014, 9, e88092. [Google Scholar] [CrossRef] [PubMed]
- Bani, L.; Orioli, V.; Pisa, G.; Fagiani, S.; Dondina, O.; Fabbri, E.; Randi, E.; Sozio, G.; Mortelliti, A. Population Genetic Structure and Sex-Biased Dispersal of the Hazel Dormouse (Muscardinus avellanarius) in a Continuous and in a Fragmented Landscape in Central Italy. Conserv. Genet. 2017, 18, 261–274. [Google Scholar] [CrossRef]
- Oklander, L.I.; Kowalewski, M.M.; Corach, D. Genetic Consequences of Habitat Fragmentation in Black-and-Gold Howler (Alouatta caraya) Populations from Northern Argentina. Int. J. Primatol. 2010, 31, 813–832. [Google Scholar] [CrossRef]
- Villaseñor, N.R.; Driscoll, D.A.; Escobar, M.A.H.; Gibbons, P.; Lindenmayer, D.B. Urbanization Impacts on Mammals across Urban-Forest Edges and a Predictive Model of Edge Effects. PLoS ONE 2014, 9, e97036. [Google Scholar] [CrossRef]
- Allen, M.; Webb, M.H.; Alves, F.; Heinsohn, R.; Stojanovic, D. Occupancy Patterns of the Introduced, Predatory Sugar Glider in Tasmanian Forests. Austral Ecol. 2018, 43, 470–475. [Google Scholar] [CrossRef]
- Gracanin, A.; Mikac, K.M. Camera Traps Reveal Overlap and Seasonal Variation in the Diel Activity of Arboreal and Semi-Arboreal Mammals. Mamm. Biol. 2022, 102, 341–355. [Google Scholar] [CrossRef]
- Potts, B. Population Genetics of Antechinus Stuartii in a Fragmented Landscape. Honours Thesis, University of Wollongong, Wollongong, Australia, 2018. [Google Scholar]
- Knipler, M.L.; Gracanin, A.; Mikac, K.M. Conservation Genomics of an Endangered Arboreal Mammal Following the 2019–2020 Australian Megafire. Sci. Rep. 2023, 13, 480. [Google Scholar] [CrossRef] [PubMed]
- Goldingay, R.L. Characteristics of Tree Hollows Used by Australian Arboreal and Scansorial Mammals. Aust. J. Zool. 2012, 59, 277–294. [Google Scholar] [CrossRef]




| Sampling Location | N | Ho | He | FIS |
|---|---|---|---|---|
| Berry East | 35 | 0.188 (0.182) | 0.200 (0.184) | 0.045 (0.203) |
| Boundary Road | 1 | 0.212 (0.409) | - | - |
| Broughton | 9 | 0.190 (0.206) | 0.202 (0.197) | 0.042 (0.304) |
| Coomonderry | 6 | 0.189 (0.228) | 0.195 (0.207) | 0.009 (0.352) |
| Escarpment | 10 | 0.202 (0.210) | 0.208 (0.194) | 0.021 (0.297) |
| SMBNP | 24 | 0.189 (0.185) | 0.202 (0.185) | 0.053 (0.228) |
| Tindalls Lane | 7 | 0.187 (0.209) | 0.207 (0.205) | 0.069 (0.341) |
| Woodhill | 2 | 0.202 (0.314) | 0.196 (0.275) | −0.126 (0.534) |
| Source of Variation | D.F. | SS | MS | % Variation | p |
|---|---|---|---|---|---|
| Between locations | 7 | 8905.74 | 1272.249 | 4.86 | 0.001 |
| Between individuals within sampling locations | 86 | 54,804.95 | 637.267 | 5.12 | 0.001 |
| Within individuals | 94 | 53,790.56 | 572.240 | 90.03 | 0.001 |
| Total | 187 | 117,501.25 | 628.349 | 100 |
| SMBNP | Broughton Creek | Boundary Road | Berry East | Woodhill | Escarpment | Coomonderry | Tindalls Lane | |
|---|---|---|---|---|---|---|---|---|
| SMBNP | - | * | * | * | * | * | * | * |
| Broughton Creek | 0.053 | - | * | * | * | * | * | * |
| Boundary Road | 0.052 | 0.044 | - | * | * | * | * | n.s. |
| Berry East | 0.048 | 0.021 | 0.055 | - | * | * | * | * |
| Woodhill | 0.073 | 0.051 | 0.069 | 0.072 | - | * | * | * |
| Escarpment | 0.064 | 0.053 | 0.042 | 0.064 | 0.057 | - | * | * |
| Coomonderry | 0.011 | 0.052 | 0.073 | 0.046 | 0.090 | 0.067 | - | * |
| Tindalls Lane | 0.057 | 0.027 | 0.005 | 0.044 | 0.043 | 0.049 | 0.057 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gracanin, A.; Knipler, M.L.; Mikac, K.M. Informing Wildlife Corridor Creation through Population Genetics of an Arboreal Marsupial in a Fragmented Landscape. Genes 2023, 14, 349. https://doi.org/10.3390/genes14020349
Gracanin A, Knipler ML, Mikac KM. Informing Wildlife Corridor Creation through Population Genetics of an Arboreal Marsupial in a Fragmented Landscape. Genes. 2023; 14(2):349. https://doi.org/10.3390/genes14020349
Chicago/Turabian StyleGracanin, Ana, Monica L. Knipler, and Katarina M. Mikac. 2023. "Informing Wildlife Corridor Creation through Population Genetics of an Arboreal Marsupial in a Fragmented Landscape" Genes 14, no. 2: 349. https://doi.org/10.3390/genes14020349
APA StyleGracanin, A., Knipler, M. L., & Mikac, K. M. (2023). Informing Wildlife Corridor Creation through Population Genetics of an Arboreal Marsupial in a Fragmented Landscape. Genes, 14(2), 349. https://doi.org/10.3390/genes14020349







