Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatment
2.2. Wnt Characterization and Manual Annotation
2.3. Phylogenetic Analysis
2.4. Differential Gene Expression (DGE)
3. Results
3.1. Wnt Genes in H. glaberrima
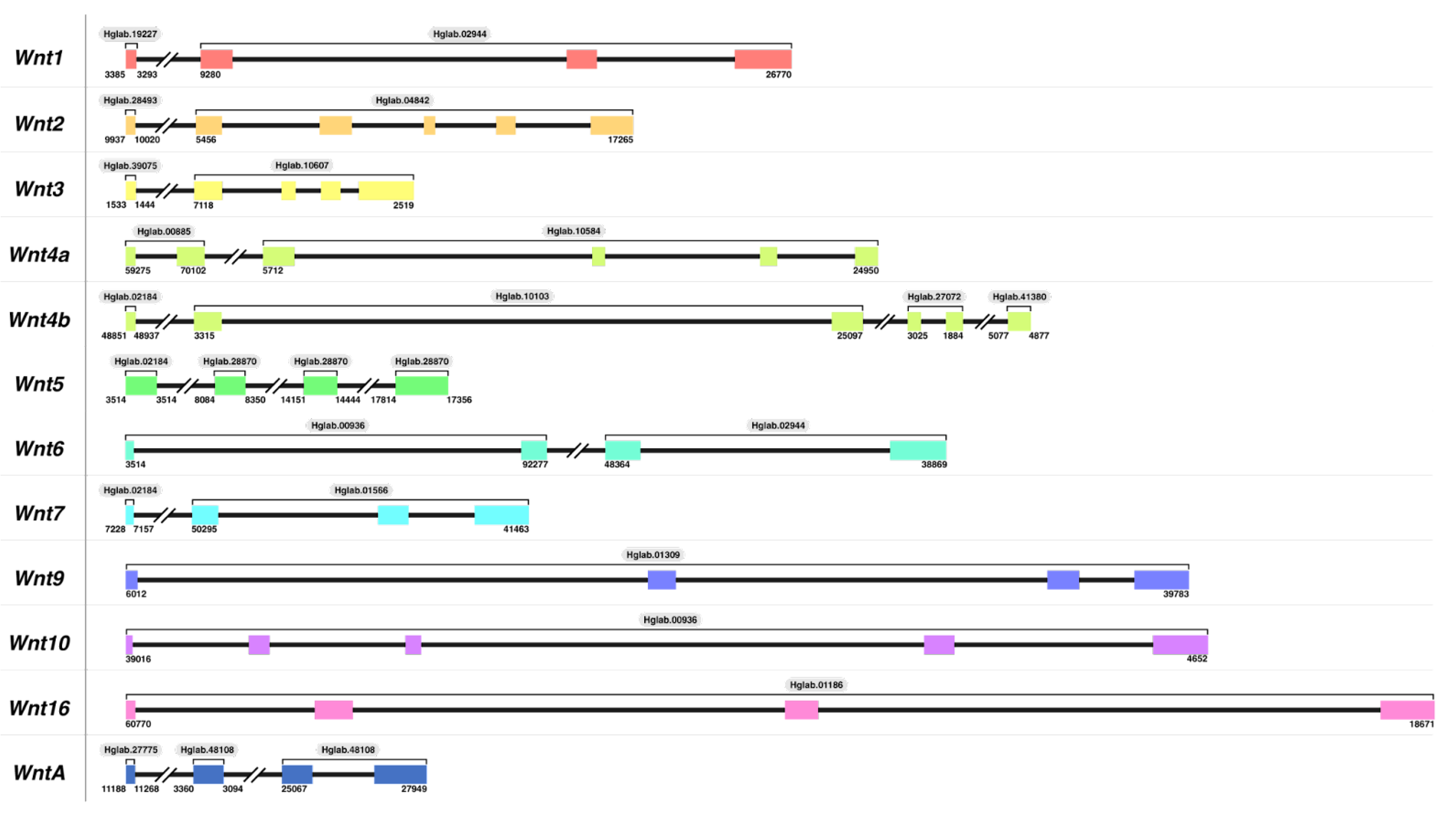
3.1.1. Wnt Gene Identification and Manual Annotation
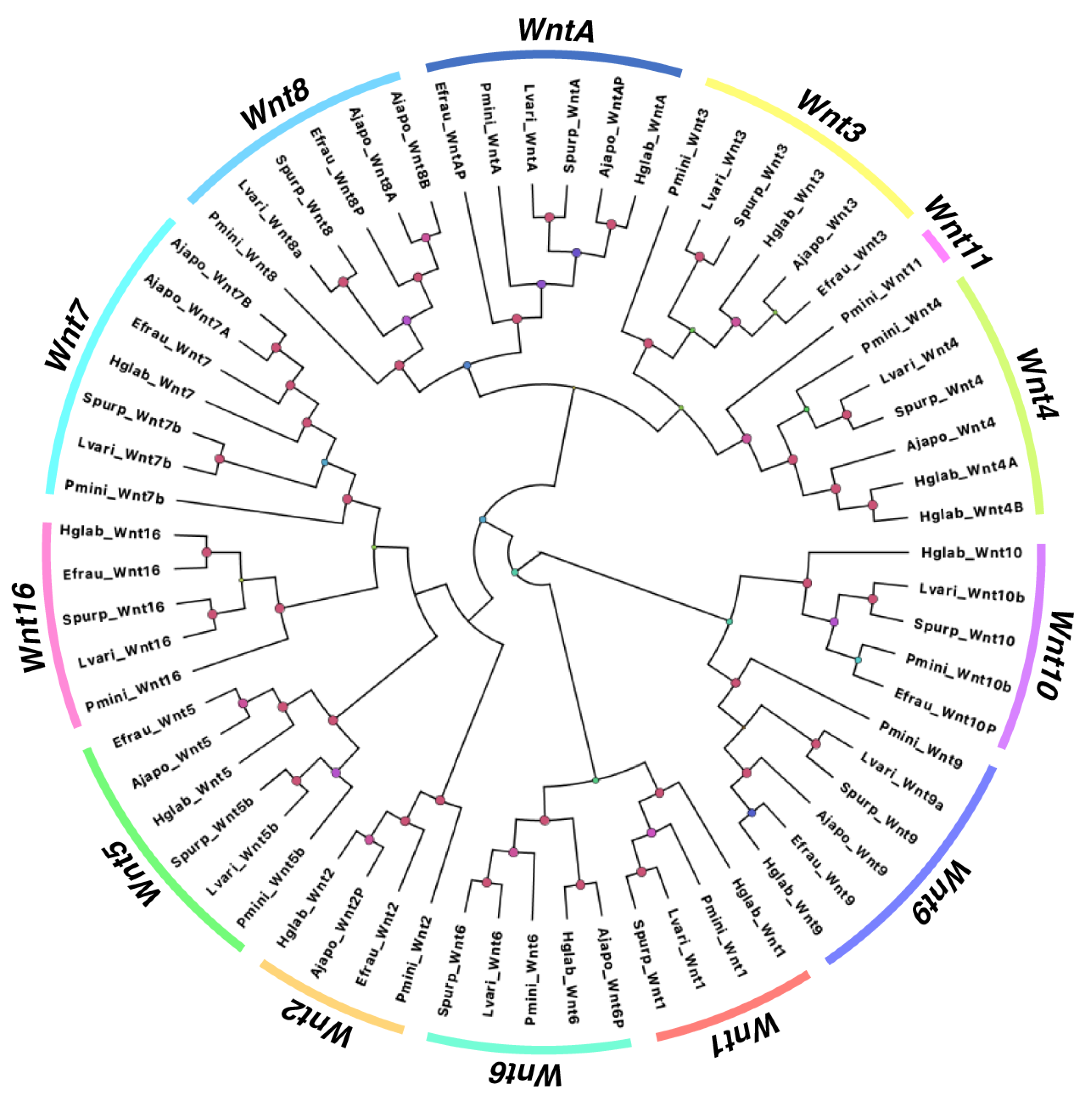
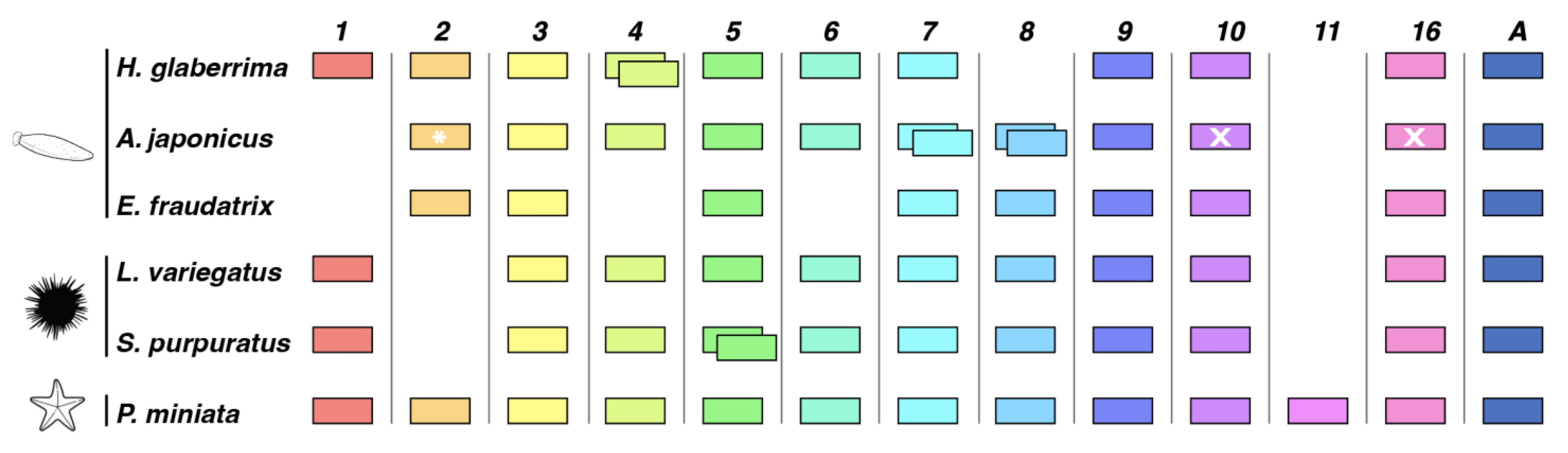
3.1.2. Comparison of Wnt Genes in Echinodermata
3.2. Expression of Wnt Signaling Genes
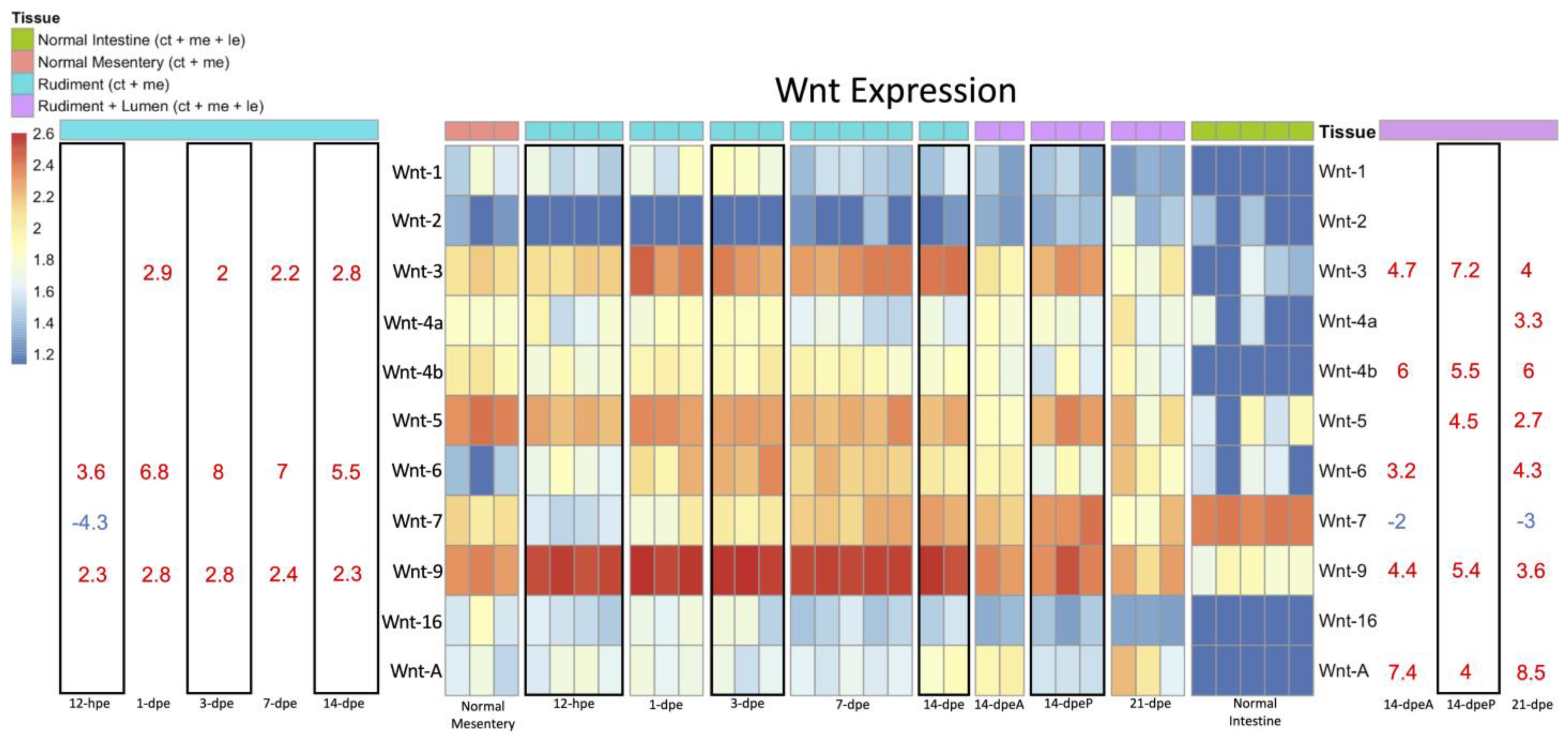
3.2.1. Wnt Expression
3.2.2. Expression of Wnt-Associated Genes
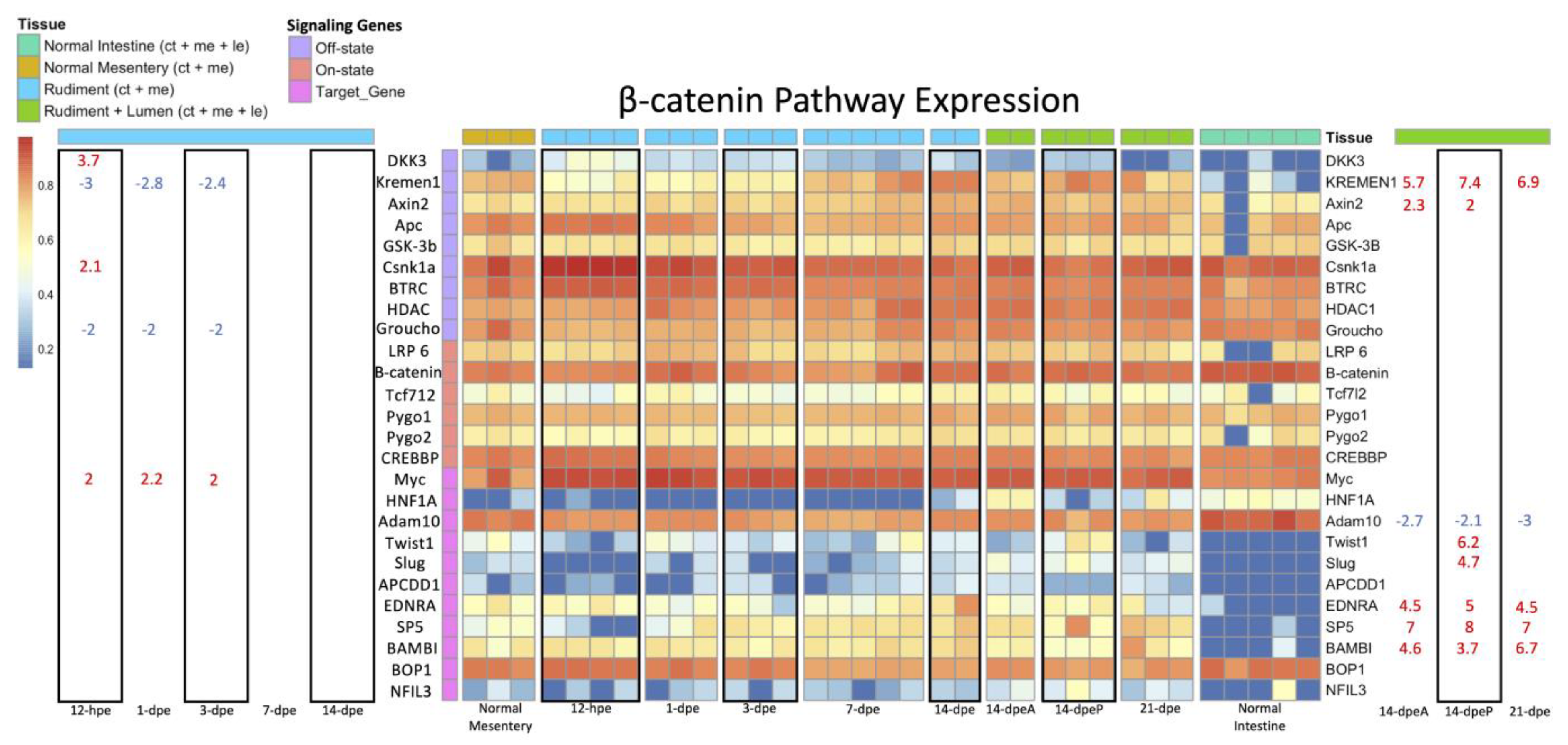
3.2.3. Wnt/β-Catenin and Wnt/PCP Pathway Gene Expression
4. Discussion
4.1. Comparison of Wnt Genes in Echinodermata
4.2. Wnt Signaling during Early-Stage Regeneration
4.3. Wnt Signaling during Late-Stage Regeneration
4.4. Wnt Signaling Pathways
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yaglova, N.V.; Tsomartova, D.A.; Obernikhin, S.S.; Nazimova, S.V. The Role of the Canonical Wnt-Signaling Pathway in Morphogenesis and Regeneration of the Adrenal Cortex in Rats Exposed to the Endocrine Disruptor Dichlorodiphenyltrichloroethane during Prenatal and Postnatal Development. Biol. Bull. Russ. Acad. Sci. 2019, 46, 74–81. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt Signal Transduction Pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Schmidt, C.; Luke, G.; Allen, S.; Valasek, P.; Muntoni, F.; Lawrence-Watt, D.; Patel, K. Canonical Wnt Signalling Induces Satellite-Cell Proliferation during Adult Skeletal Muscle Regeneration. J. Cell Sci. 2008, 121, 2939–2950. [Google Scholar] [CrossRef] [PubMed]
- He, T.-C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of C-MYC as a Target of the APC Pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Tetsu, O.; McCormick, F. B-Catenin Regulates Expression of Cyclin D1 in Colon Carcinoma Cells. Nature 1999, 398, 422–426. [Google Scholar] [CrossRef]
- Kim, J.-S.; Crooks, H.; Dracheva, T.; Nishanian, T.G.; Singh, B.; Jen, J.; Waldman, T. Oncogenic B-Catenin Is Required for Bone Morphogenetic Protein 4 Expression in Human Cancer Cells. Cancer Res. 2002, 62, 2744–2748. [Google Scholar]
- Wu, X.; Tu, X.; Joeng, K.S.; Hilton, M.J.; Williams, D.A.; Long, F. Rac1 Activation Controls Nuclear Localization of β-Catenin during Canonical Wnt Signaling. Cell 2008, 133, 340–353. [Google Scholar] [CrossRef]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-Kinase/ROCK: A Key Regulator of the Cytoskeleton and Cell Polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Golenia, G.; Gatie, M.I.; Kelly, G.M. Frizzled Gene Expression and Negative Regulation of Canonical WNT–β-Catenin Signaling in Mouse F9 Teratocarcinoma Cells. Biochem. Cell Biol. 2017, 95, 251–262. [Google Scholar] [CrossRef]
- Shi, D.-L. Planar Cell Polarity Regulators in Asymmetric Organogenesis during Development and Disease. J. Genet. Genom. 2022, in press. [Google Scholar] [CrossRef]
- Mentink, R.A.; Rella, L.; Radaszkiewicz, T.W.; Gybel, T.; Betist, M.C.; Bryja, V.; Korswagen, H.C. The Planar Cell Polarity Protein VANG-1/Vangl Negatively Regulates Wnt/β-Catenin Signaling through a Dvl Dependent Mechanism. PLoS Genet. 2018, 14, e1007840. [Google Scholar] [CrossRef] [PubMed]
- Tian, A.; Benchabane, H.; Ahmed, Y. Wingless/Wnt Signaling in Intestinal Development, Homeostasis, Regeneration and Tumorigenesis: A Drosophila Perspective. JDB 2018, 6, 8. [Google Scholar] [CrossRef]
- Perochon, J.; Carroll, L.; Cordero, J. Wnt Signalling in Intestinal Stem Cells: Lessons from Mice and Flies. Genes 2018, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Ouladan, S.; Gregorieff, A. Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation. IJMS 2021, 22, 7043. [Google Scholar] [CrossRef]
- Mao, J.; Fan, S.; Ma, W.; Fan, P.; Wang, B.; Zhang, J.; Wang, H.; Tang, B.; Zhang, Q.; Yu, X.; et al. Roles of Wnt/β-Catenin Signaling in the Gastric Cancer Stem Cells Proliferation and Salinomycin Treatment. Cell Death Dis. 2014, 5, e1039. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.N.; Kim, M.J.; Jung, Y.-S.; Lien, E.M.; Jun, S.; Park, J.-I. Quiescence Exit of Tert+ Stem Cells by Wnt/β-Catenin Is Indispensable for Intestinal Regeneration. Cell Rep. 2017, 21, 2571–2584. [Google Scholar] [CrossRef]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant Sources of Wnt Regulate Intestinal Stem Cells and Promote Formation of Paneth Cells. Gastroenterology 2012, 143, 1518–1529.e7. [Google Scholar] [CrossRef]
- Miyoshi, H.; Ajima, R.; Luo, C.T.; Yamaguchi, T.P.; Stappenbeck, T.S. Wnt5a Potentiates TGF-β Signaling to Promote Colonic Crypt Regeneration After Tissue Injury. Science 2012, 338, 108–113. [Google Scholar] [CrossRef]
- Sigal, M.; Logan, C.Y.; Kapalczynska, M.; Mollenkopf, H.-J.; Berger, H.; Wiedenmann, B.; Nusse, R.; Amieva, M.R.; Meyer, T.F. Stromal R-Spondin Orchestrates Gastric Epithelial Stem Cells and Gland Homeostasis. Nature 2017, 548, 451–455. [Google Scholar] [CrossRef]
- Gregorieff, A.; Pinto, D.; Begthel, H.; Destrée, O.; Kielman, M.; Clevers, H. Expression Pattern of Wnt Signaling Components in the Adult Intestine. Gastroenterology 2005, 129, 626–638. [Google Scholar] [CrossRef]
- Lin, G.; Xu, N.; Xi, R. Paracrine Wingless Signalling Controls Self-Renewal of Drosophila Intestinal Stem Cells. Nature 2008, 455, 1119–1123. [Google Scholar] [CrossRef]
- Buchon, N.; Osman, D.; David, F.P.A.; Yu Fang, H.; Boquete, J.-P.; Deplancke, B.; Lemaitre, B. Morphological and Molecular Characterization of Adult Midgut Compartmentalization in Drosophila. Cell Rep. 2013, 3, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, A.; Benchabane, H.; Tacchelly-Benites, O.; Yang, E.; Nojima, H.; Ahmed, Y. The ADP-Ribose Polymerase Tankyrase Regulates Adult Intestinal Stem Cell Proliferation during Homeostasis in Drosophila. Development 2016, 143, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Tian, A.; Benchabane, H.; Wang, Z.; Ahmed, Y. Regulation of Stem Cell Proliferation and Cell Fate Specification by Wingless/Wnt Signaling Gradients Enriched at Adult Intestinal Compartment Boundaries. PLoS Genet. 2016, 12, e1005822. [Google Scholar] [CrossRef]
- Quispe-Parra, D.; Valentín, G.; García-Arrarás, J.E. A Roadmap for Intestinal Regeneration. Int. J. Dev. Biol. 2021, 52, 427–437. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Estrada-Rodgers, L.; Santiago, R.; Torres, I.I.; Díaz-Miranda, L.; Torres-Avillán, I. Cellular Mechanisms of Intestine Regeneration in the Sea Cucumber, Holothuria Glaberrima Selenka (Holothuroidea:Echinodermata). J. Exp. Zool. 1998, 281, 288–304. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Valentín-Tirado, G.; Flores, J.E.; Rosa, R.J.; Rivera-Cruz, A.; San Miguel-Ruiz, J.E.; Tossas, K. Cell Dedifferentiation and Epithelial to Mesenchymal Transitions during Intestinal Regeneration in H. Glaberrima. BMC Dev. Biol. 2011, 11, 61. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Bello, S.A.; Malavez, S. The Mesentery as the Epicenter for Intestinal Regeneration. Semin. Cell Dev. Biol. 2019, 92, 45–54. [Google Scholar] [CrossRef]
- Medina-Feliciano, J.G.; García-Arrarás, J.E. Regeneration in Echinoderms: Molecular Advancements. Front. Cell Dev. Biol. 2021, 9, 768641. [Google Scholar] [CrossRef]
- Ortiz-Pineda, P.A.; Ramírez-Gómez, F.; Pérez-Ortiz, J.; González-Díaz, S.; Santiago-De Jesús, F.; Hernández-Pasos, J.; Del Valle-Avila, C.; Rojas-Cartagena, C.; Suárez-Castillo, E.C.; Tossas, K.; et al. Gene Expression Profiling of Intestinal Regeneration in the Sea Cucumber. BMC Genom. 2009, 10, 262. [Google Scholar] [CrossRef]
- Mashanov, V.S.; Zueva, O.R.; Garcia-Arraras, J.E. Expression of Wnt9, TCTP, and Bmp1/Tll in Sea Cucumber Visceral Regeneration. Gene Expr. Patterns 2012, 12, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Parra, D.J.; Medina-Feliciano, J.G.; Cruz-González, S.; Ortiz-Zuazaga, H.; García-Arrarás, J.E. Transcriptomic Analysis of Early Stages of Intestinal Regeneration in Holothuria Glaberrima. Sci. Rep. 2021, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Alicea-Delgado, M.; García-Arrarás, J.E. Wnt/β-Catenin Signaling Pathway Regulates Cell Proliferation but Not Muscle Dedifferentiation nor Apoptosis during Sea Cucumber Intestinal Regeneration. Dev. Biol. 2021, 480, 105–113. [Google Scholar] [CrossRef]
- Li, X.; Sun, L.; Yang, H.; Zhang, L.; Miao, T.; Xing, L.; Huo, D. Identification and Expression Characterization of WntA during Intestinal Regeneration in the Sea Cucumber Apostichopus japonicus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 210, 55–63. [Google Scholar] [CrossRef]
- Sun, L.N.; Yang, H.S.; Chen, M.Y.; Xu, D.X. Cloning and Expression Analysis of Wnt6 and Hox6 during Intestinal Regeneration in the Sea Cucumber Apostichopus japonicus. Genet. Mol. Res. 2013, 12, 5321–5334. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, L.; Yuan, J.; Sun, Y.; Gao, Y.; Zhang, L.; Li, S.; Dai, H.; Hamel, J.-F.; Liu, C.; et al. The Sea Cucumber Genome Provides Insights into Morphological Evolution and Visceral Regeneration. PLoS Biol. 2017, 15, e2003790. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Gao, Y.; Sun, L.; Jin, S.; Zhang, X.; Liu, C.; Li, F.; Xiang, J. Wnt Signaling Pathway Linked to Intestinal Regeneration via Evolutionary Patterns and Gene Expression in the Sea Cucumber Apostichopus japonicus. Front. Genet. 2019, 10, 112. [Google Scholar] [CrossRef]
- Girich, A.S.; Isaeva, M.P.; Dolmatov, I.Y. Wnt and Frizzled Expression during Regeneration of Internal Organs in the Holothurian Eupentacta Fraudatrix: Wnt and Frizzled in Holothurian Regeneration. Wound Rep. and Reg. 2017, 25, 828–835. [Google Scholar] [CrossRef]
- Medina-Feliciano, J.G.; Pirro, S.; García-Arrarás, J.E.; Mashanov, V.; Ryan, J.F. Draft Genome of the Sea Cucumber Holothuria Glaberrima, a Model for the Study of Regeneration. Front. Mar. Sci. 2021, 8, 603410. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling Differential Gene Expression Analysis for de Novoassembled Transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Vosburg, C.; Reynolds, M.; Noel, R.; Shippy, T.; Hosmani, P.S.; Flores-Gonzalez, M.; Mueller, L.A.; Hunter, W.B.; Brown, S.J.; D’Elia, T.; et al. Utilizing a Chromosomal-Length Genome Assembly to Annotate the Wnt Signaling Pathway in the Asian Citrus Psyllid, Diaphorina citri. Gigabyte 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Molven, A.; Njølstad, P.R.; Fjose, A. Genomic Structure and Restricted Neural Expression of the Zebrafish Wnt-1 (Int-1) Gene. EMBO J. 1991, 10, 799–807. [Google Scholar] [CrossRef]
- Miller, J.R. The wnts. Gen. Biol. 2001, 3, 1–15. [Google Scholar] [CrossRef]
- Girich, A.S.; Boyko, A.V. Wnt and Frizzled Genes in Echinoderms. Russ. J. Mar. Biol. 2019, 45, 302–312. [Google Scholar] [CrossRef]
- Bello, S.A.; Torres-Gutiérrez, V.; Rodríguez-Flores, E.J.; Toledo-Román, E.J.; Rodríguez, N.; Díaz-Díaz, L.M.; Vázquez-Figueroa, L.D.; Cuesta, J.M.; Grillo-Alvarado, V.; Amador, A.; et al. Insights into Intestinal Regeneration Signaling Mechanisms. Dev. Biol. 2020, 458, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Yu, H. Wnt6 Contributes Tumorigenesis and Development of Colon Cancer via Its Effects on Cell Proliferation, Apoptosis, Cell-cycle and Migration. Oncol. Lett. 2018, 16, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Benhaj, K.; Akcali, K.; Ozturk, M. Redundant Expression of Canonical Wnt Ligands in Human Breast Cancer Cell Lines. Oncol. Rep. 2006, 15, 701–707. [Google Scholar] [CrossRef]
- Matsu-ura, T.; Dovzhenok, A.; Aihara, E.; Rood, J.; Le, H.; Ren, Y.; Rosselot, A.E.; Zhang, T.; Lee, C.; Obrietan, K.; et al. Intercellular Coupling of the Cell Cycle and Circadian Clock in Adult Stem Cell Culture. Mol. Cell 2016, 64, 900–912. [Google Scholar] [CrossRef]
- Wu, Y.; Ginther, C.; Kim, J.; Mosher, N.; Chung, S.; Slamon, D.; Vadgama, J.V. Expression of Wnt3 Activates Wnt/β-Catenin Pathway and Promotes EMT-like Phenotype in Trastuzumab-Resistant HER2-Overexpressing Breast Cancer Cells. Mol. Cancer Res. 2012, 10, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Baulies, A.; Angelis, N.; Li, V.S.W. Hallmarks of Intestinal Stem Cells. Development 2020, 147, dev182675. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, D.J.; Phesse, T.J.; Barker, N.; Schwab, R.H.M.; Amin, N.; Malaterre, J.; Stange, D.E.; Nowell, C.J.; Currie, S.A.; Saw, J.T.S.; et al. Frizzled7 Functions as a Wnt Receptor in Intestinal Epithelial Lgr5+ Stem Cells. Stem Cell Rep. 2015, 4, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Mashanov, V.S.; Zueva, O.R.; Rojas-Catagena, C.; Garcia-Arraras, J.E. Visceral Regeneration in a Sea Cucumber Involves Extensive Expression of Survivin and Mortalin Homologs in the Mesothelium. BMC Dev. Biol. 2010, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.R.; Willems, E.; Wawrzak, D.; Hendrickx, M.; Agbor Agbor, T.; Leyns, L. Expression of Frizzled5, Frizzled7, and Frizzled10 during Early Mouse Development and Interactions with Canonical Wnt Signaling. Dev. Dyn. 2007, 236, 2011–2019. [Google Scholar] [CrossRef]
- Janssen, R.; Pechmann, M.; Turetzek, N. A Chelicerate Wnt Gene Expression Atlas: Novel Insights into the Complexity of Arthropod Wnt-Patterning. EvoDevo 2021, 12, 12. [Google Scholar] [CrossRef]
- Andre, P.; Song, H.; Kim, W.; Kispert, A.; Yang, Y. Wnt5a and Wnt11 Regulate Mammalian Anterior-Posterior Axis Elongation. Development 2015, 142, 1516–1527. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth Cells Constitute the Niche for Lgr5 Stem Cells in Intestinal Crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef]
- Van Es, J.H.; Jay, P.; Gregorieff, A.; van Gijn, M.E.; Jonkheer, S.; Hatzis, P.; Thiele, A.; van den Born, M.; Begthel, H.; Brabletz, T.; et al. Wnt Signalling Induces Maturation of Paneth Cells in Intestinal Crypts. Nat. Cell Biol. 2005, 7, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Krawetz, R.; Kelly, G.M. Wnt6 Induces the Specification and Epithelialization of F9 Embryonal Carcinoma Cells to Primitive Endoderm. Cell. Signal. 2008, 20, 506–517. [Google Scholar] [CrossRef]
- Jin, Y.-R.; Han, X.H.; Nishimori, K.; Ben-Avraham, D.; Oh, Y.J.; Shim, J.; Yoon, J.K. Canonical WNT/β-Catenin Signaling Activated by WNT9b and RSPO2 Cooperation Regulates Facial Morphogenesis in Mice. Front. Cell Dev. Biol. 2020, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Dush, M.K.; Nascone-Yoder, N.M. Vangl2 Coordinates Cell Rearrangements during Gut Elongation. Dev. Dyn. 2019, 248, 569–582. [Google Scholar] [CrossRef]
- Matsuyama, M.; Aizawa, S.; Shimono, A. Sfrp Controls Apicobasal Polarity and Oriented Cell Division in Developing Gut Epithelium. PLoS Genet. 2009, 5, e1000427. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Auger, N.A.; Medina-Feliciano, J.G.; Quispe-Parra, D.J.; Colón-Marrero, S.; Ortiz-Zuazaga, H.; García-Arrarás, J.E. Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis. Genes 2023, 14, 309. https://doi.org/10.3390/genes14020309
Auger NA, Medina-Feliciano JG, Quispe-Parra DJ, Colón-Marrero S, Ortiz-Zuazaga H, García-Arrarás JE. Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis. Genes. 2023; 14(2):309. https://doi.org/10.3390/genes14020309
Chicago/Turabian StyleAuger, Noah A., Joshua G. Medina-Feliciano, David J. Quispe-Parra, Stephanie Colón-Marrero, Humberto Ortiz-Zuazaga, and José E. García-Arrarás. 2023. "Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis" Genes 14, no. 2: 309. https://doi.org/10.3390/genes14020309
APA StyleAuger, N. A., Medina-Feliciano, J. G., Quispe-Parra, D. J., Colón-Marrero, S., Ortiz-Zuazaga, H., & García-Arrarás, J. E. (2023). Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis. Genes, 14(2), 309. https://doi.org/10.3390/genes14020309





