Omics Data Preprocessing for Machine Learning: A Case Study in Childhood Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of Case Study Population and Data
2.2. Data Pre-Processing Guidelines and Analytical Assessment of ML Predictive Models
2.2.1. GWAS Data
2.2.2. EWAS Data
2.2.3. Biochemistry, Anthropometrical, and Clinical Data
2.3. Basis and Recommendations That Must Guide the Selection of a ML Algorithm and the Experimental Design
2.3.1. Experimental Design
2.3.2. Selection of ML Algorithms and Classification Metrics
2.3.3. SHAP Explanations
3. Results
4. Discussion
4.1. Main Challenges That Are Usually Faced in Omics ML Predictive Modeling
4.2. Analysis of ML Results and Insights from the Case Study
- Models trained using the imbalanced datasets show better sensitivity at the expense of very poor specificity, while datasets balanced during the training stage provide more consistent values for both metrics and greater generalizability to unseen data of any kind.
- When the training dataset is balanced, the biochemistry dataset provides the best results in terms of F1, G-mean, accuracy, sensitivity, and specificity, followed by EWAS and GWAS; this leads us to conclude that combining biochemistry and EWAS datasets may be a promising strategy to improve these results. As Table 2 shows, the classifiers generated by OneR and CART obtain slightly higher values for the metrics analyzed on the biochemical datasets. However, XGBoost obtains similar results for the omics and higher values for the other two omics, presenting robust behavior in all of them [36].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goecks, J.; Jalili, V.; Heiser, L.M.; Gray, J.W. How Machine Learning Will Transform Biomedicine. Cell 2020, 181, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized nutrition by prediction of glycemic responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Sammut, S.J.; Crispin-Ortuzar, M.; Chin, S.F.; Provenzano, E.; Bardwell, H.A.; Ma, W.; Cope, W.; Dariush, A.; Dawson, S.J.; Abraham, J.E.; et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 2022, 601, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, L.; Xu, Y.; Yang, J. Machine learning meets omics: Applications and perspectives. Briefings Bioinform. 2021, 23, bbab460. [Google Scholar] [CrossRef] [PubMed]
- Whalen, S.; Schreiber, J.; Noble, W.S.; Pollard, K.S. Navigating the pitfalls of applying machine learning in genomics. Nat. Rev. Genet. 2022, 23, 169–181. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Riley, R.D.; Snell, K.I.E.; Martin, G.P.; Whittle, R.; Archer, L.; Sperrin, M.; Collins, G.S. Penalization and shrinkage methods produced unreliable clinical prediction models especially when sample size was small. J. Clin. Epidemiol. 2021, 132, 88–96. [Google Scholar] [CrossRef]
- Yang, P.; Huang, H.; Liu, C. Feature selection revisited in the single-cell era. Genome Biol. 2021, 22, 321. [Google Scholar] [CrossRef]
- Torres-Martos, Á.; Anguita-Ruiz, A.; Bustos-Aibar, M.; Cámara-Sánchez, S.; Alcalá, R.; Aguilera, C.M.; Alcalá-Fdez, J. Human Multi-omics Data Pre-processing for Predictive Purposes Using Machine Learning: A Case Study in Childhood Obesity. In Proceedings of the Bioinformatics and Biomedical Engineering, Gran Canaria, Spain, 27–30 June 2022; Rojas, I., Valenzuela, O., Rojas, F., Herrera, L.J., Ortuño, F., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 359–374. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Herrera, F.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Anguita Ruiz, A.M. Multi-Omics Integration and Machine Learning for the Identification of Molecular Markers of Insulin Resistance in Prepubertal and Pubertal Children with Obesity. Ph.D. Thesis, University of Granada, Granada, Spain, 2021. [Google Scholar]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulou, K.; Walter, K. Quality Control of Common and Rare Variants. In Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 25–36. [Google Scholar] [CrossRef]
- Phocas, F. Genotyping, the usefulness of imputation to increase SNP density, and imputation methods and tools. In Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 113–138. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Sheikhali Babaei, M.; Sharp, G.; Hatcher, C.; Vega-Salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A database of epigenome-wide association studies. Wellcome Open Res. 2022, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Oostra, B.A.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Lotta, L.A.; Gulati, P.; Day, F.R.; Payne, F.; Ongen, H.; van de Bunt, M.; Gaulton, K.J.; Eicher, J.D.; Sharp, S.J.; Luan, J.; et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat. Genet. 2017, 49, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, P.; Knapič, E.; Kokošar, J.; Kovač, J.; Jerala, R.; Battelino, T.; Horvat, S. Identification of novel alleles associated with insulin resistance in childhood obesity using pooled-DNA genome-wide association study approach. Int. J. Obes. 2018, 42, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Maksimovic, J.; Phipson, B.; Oshlack, A. A cross-package Bioconductor workflow for analysing methylation array data. F1000Research 2016, 5, 1281. [Google Scholar] [CrossRef]
- Anguita-Ruiz, A.; Torres-Martos, A.; Ruiz-Ojeda, F.; Alcalá-Fdez, J.; Bueno, G.; Gil-Campos, M.; Roa-Rivas, J.; Moreno, L.; Gil, A.; Leis, R.; et al. Integrative analysis of blood cells DNA methylation, transcriptomics and genomics identifies novel epigenetic regulatory mechanisms of insulin resistance during puberty in children with obesity. medRxiv 2022, 1–70. [Google Scholar] [CrossRef]
- Van Buuren, S. Multiple imputation of discrete and continuous data by fully conditional specification. Stat. Methods Med. Res. 2007, 16, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Stekhoven, D.J.; Bühlmann, P. MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. Resampling Methods. In An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2013; pp. 175–201. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Hvitfeldt, E. Themis: Extra Recipes Steps for Dealing with Unbalanced Data, R Package Version 0.1.0; CRAN: Los Angeles, CA, USA, 2020. Available online: https://CRAN.R-project.org/package=themis (accessed on 16 December 2022).
- Fernandez, A.; Garcia, S.; Galar, M.; Prati, R.C.; Krawczyk, B.; Herrera, F. Performance Measures. In Learning from Imbalanced Data Sets; Springer Cham: Basel, Switzerland, 2018; pp. 47–61. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4793–4813. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2017; Volume 30, pp. 4768–4777. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Holte, R.C. Very Simple Classification Rules Perform Well on Most Commonly Used Datasets. Mach. Learn. 1993, 11, 63–903. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Routledge: New York, NY, USA, 1984. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Fortin, J.P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.T.; Hansen, K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef]
- Martínez-Uña, M.; López-Mancheño, Y.; Diéguez, C.; Fernández-Rojo, M.A.; Novelle, M.G. Unraveling the role of leptin in liver function and its relationship with liver diseases. Int. J. Mol. Sci. 2020, 21, 9368. [Google Scholar] [CrossRef]
- Ardestani, A.; Lupse, B.; Maedler, K. Hippo Signaling: Key Emerging Pathway in Cellular and Whole-Body Metabolism. Trends Endocrinol. Metab. 2018, 29, 492–509. [Google Scholar] [CrossRef] [PubMed]

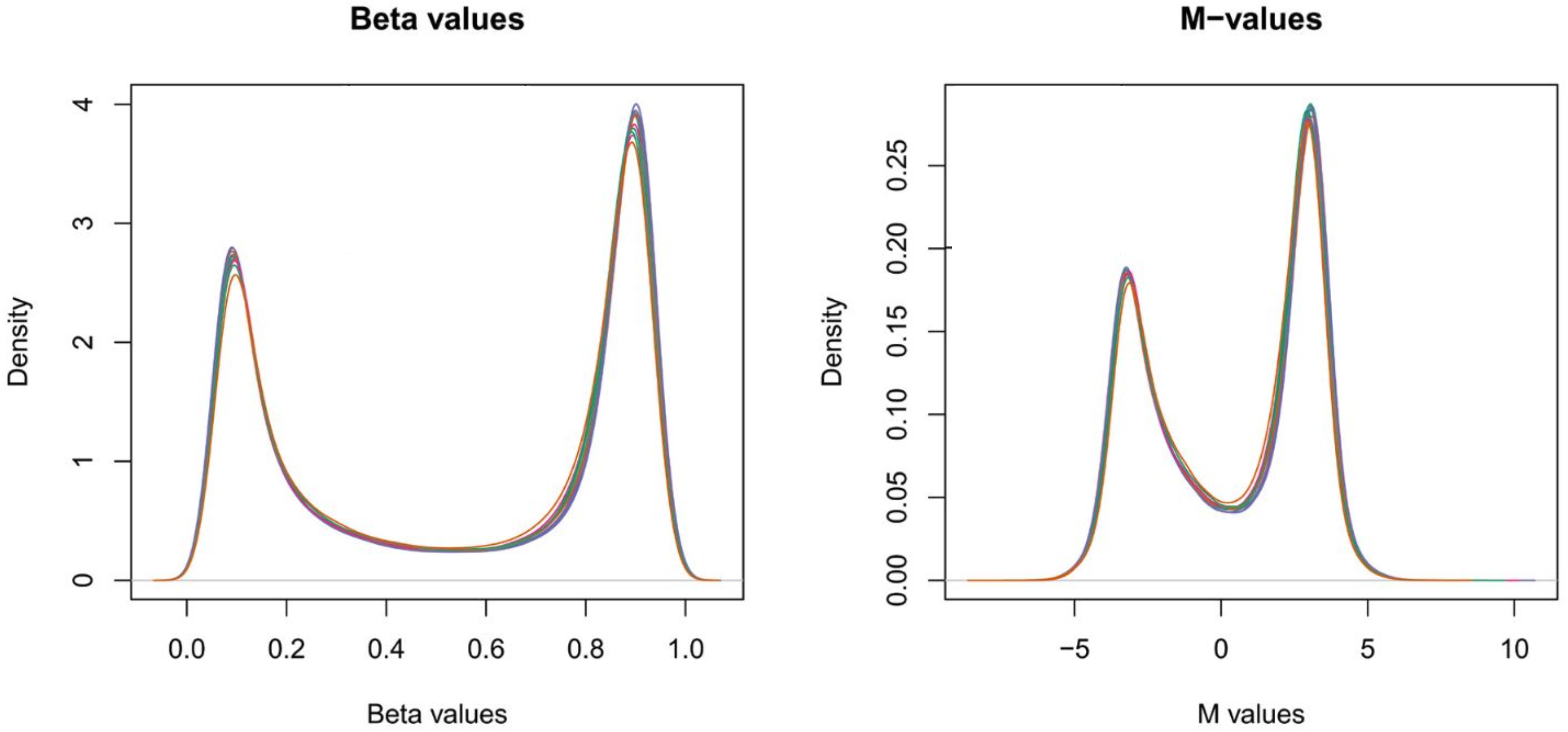

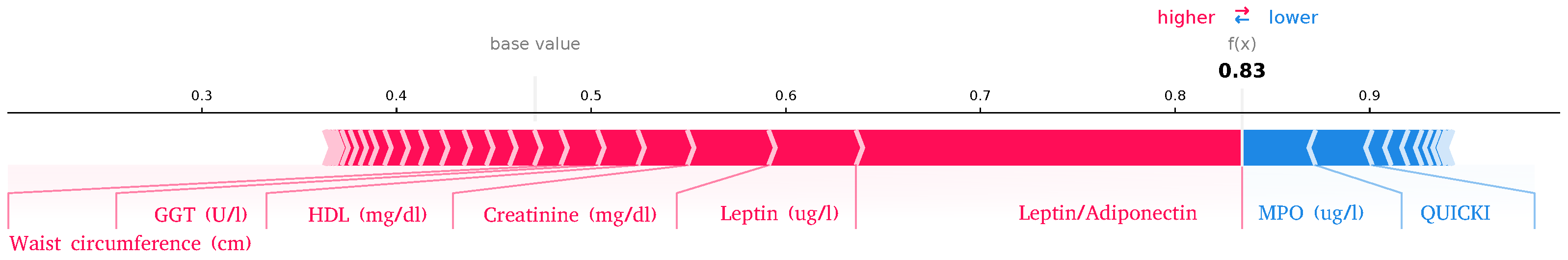
| GWAS | EWAS | Biochemistry | |
|---|---|---|---|
| Initial variables | 651,563 | 866,091 | 48 |
| Variables with low quality or missing values | 138,626 | 31,184 | 14 |
| (21.27%) | (3.60%) | (15.2%) | |
| % missing values after quality filtering | 0% | 0% | 0.9% |
| Final number of variables | 5,894,726 | 834,371 | 34 |
| Final number of variables after feature selection | 151 | 267 | 34 |
| OneR | Datasets | Datasets (Undersampling) | ||||
|---|---|---|---|---|---|---|
| Metrics | GWAS | EWAS | Biochem. | GWAS | EWAS | Biochem. |
| G-mean | 0.27 | 0.44 | 0.46 | 0.40 | 0.44 | 0.67 |
| AUC | 0.48 | 0.51 | 0.54 | 0.42 | 0.44 | 0.67 |
| F1 | 0.78 | 0.73 | 0.78 | 0.48 | 0.43 | 0.66 |
| Accuracy | 0.65 | 0.62 | 0.67 | 0.42 | 0.44 | 0.67 |
| Sensitivity | 0.88 | 0.76 | 0.83 | 0.55 | 0.45 | 0.62 |
| Specificity | 0.08 | 0.26 | 0.25 | 0.29 | 0.42 | 0.73 |
| CART | datasets | datasets (undersampling) | ||||
| Metrics | GWAS | EWAS | Biochem. | GWAS | EWAS | Biochem. |
| G-mean | 0.00 | 0.44 | 0.09 | 0.41 | 0.52 | 0.66 |
| AUC | 0.50 | 0.52 | 0.49 | 0.47 | 0.53 | 0.67 |
| F1 | 0.83 | 0.75 | 0.82 | 0.55 | 0.52 | 0.62 |
| Accuracy | 0.71 | 0.63 | 0.69 | 0.47 | 0.51 | 0.67 |
| Sensitivity | 1.00 | 0.79 | 0.97 | 0.70 | 0.55 | 0.58 |
| Specificity | 0.00 | 0.24 | 0.01 | 0.23 | 0.48 | 0.76 |
| XGBoost | datasets | datasets (undersampling) | ||||
| Metrics | GWAS | EWAS | Biochem. | GWAS | EWAS | Biochem. |
| G-mean | 0.53 | 0.48 | 0.44 | 0.60 | 0.62 | 0.64 |
| AUC | 0.65 | 0.67 | 0.59 | 0.65 | 0.70 | 0.66 |
| F1 | 0.79 | 0.82 | 0.74 | 0.59 | 0.59 | 0.64 |
| Accuracy | 0.69 | 0.72 | 0.62 | 0.60 | 0.62 | 0.64 |
| Sensitivity | 0.82 | 0.91 | 0.77 | 0.61 | 0.59 | 0.62 |
| Specificity | 0.35 | 0.25 | 0.25 | 0.59 | 0.64 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Martos, Á.; Bustos-Aibar, M.; Ramírez-Mena, A.; Cámara-Sánchez, S.; Anguita-Ruiz, A.; Alcalá, R.; Aguilera, C.M.; Alcalá-Fdez, J. Omics Data Preprocessing for Machine Learning: A Case Study in Childhood Obesity. Genes 2023, 14, 248. https://doi.org/10.3390/genes14020248
Torres-Martos Á, Bustos-Aibar M, Ramírez-Mena A, Cámara-Sánchez S, Anguita-Ruiz A, Alcalá R, Aguilera CM, Alcalá-Fdez J. Omics Data Preprocessing for Machine Learning: A Case Study in Childhood Obesity. Genes. 2023; 14(2):248. https://doi.org/10.3390/genes14020248
Chicago/Turabian StyleTorres-Martos, Álvaro, Mireia Bustos-Aibar, Alberto Ramírez-Mena, Sofía Cámara-Sánchez, Augusto Anguita-Ruiz, Rafael Alcalá, Concepción M. Aguilera, and Jesús Alcalá-Fdez. 2023. "Omics Data Preprocessing for Machine Learning: A Case Study in Childhood Obesity" Genes 14, no. 2: 248. https://doi.org/10.3390/genes14020248
APA StyleTorres-Martos, Á., Bustos-Aibar, M., Ramírez-Mena, A., Cámara-Sánchez, S., Anguita-Ruiz, A., Alcalá, R., Aguilera, C. M., & Alcalá-Fdez, J. (2023). Omics Data Preprocessing for Machine Learning: A Case Study in Childhood Obesity. Genes, 14(2), 248. https://doi.org/10.3390/genes14020248












