Genome-Wide Association Study to Identify Possible Candidate Genes of Snap Bean Leaf and Pod Color
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Data Acquisition
2.3. Genome-Wide Association Study
2.4. Candidate Gene Research
3. Results
3.1. Narrow-Sense Heritability
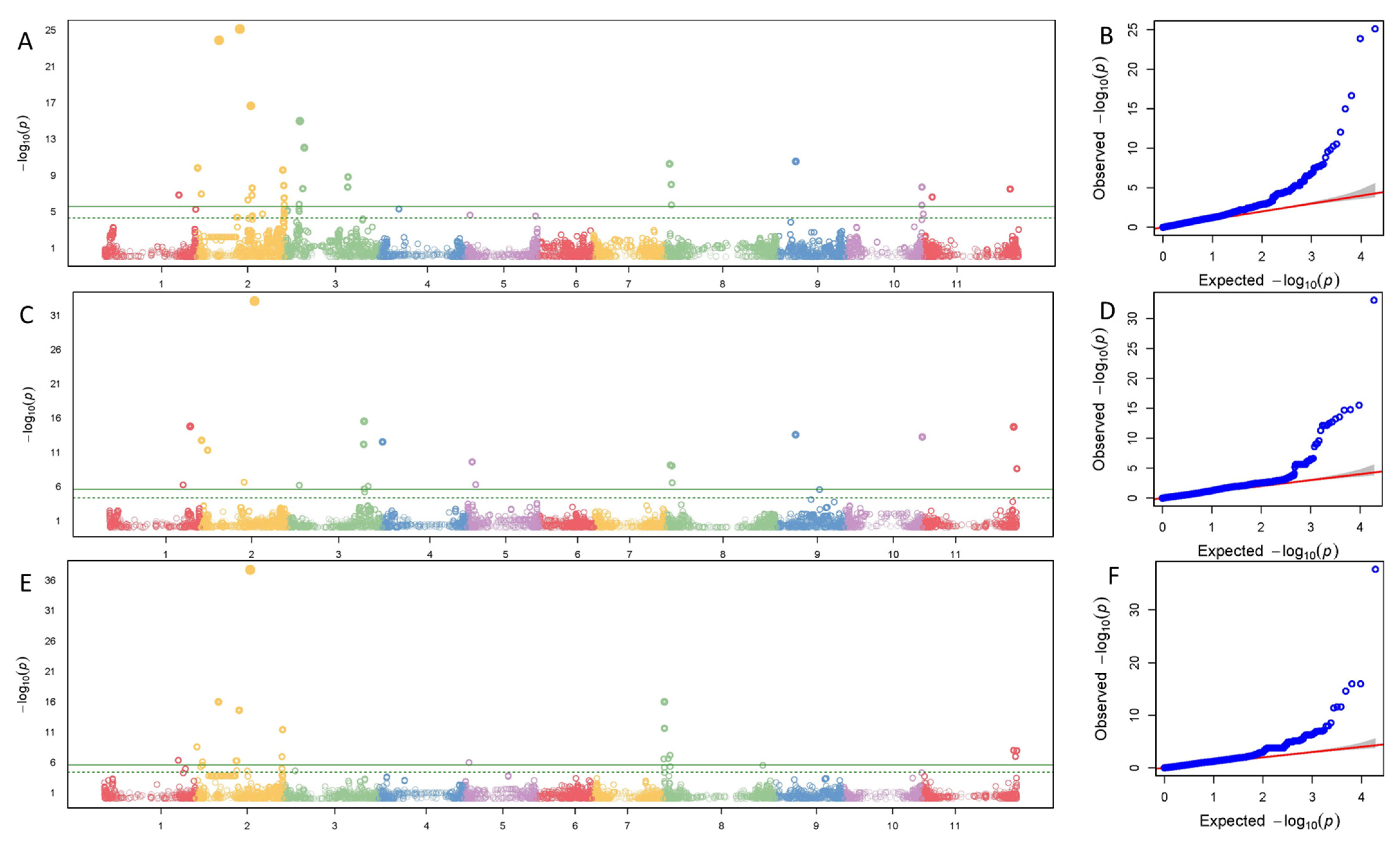
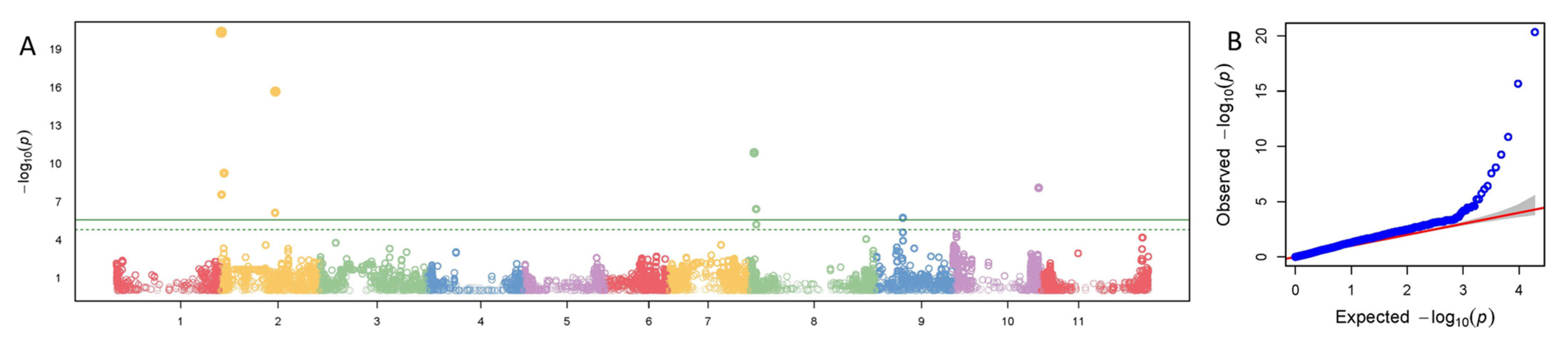
3.2. Significant SNPs
3.2.1. Overall Description of Significant SNPs
3.2.2. SNPs Associated with More than One Tissue
3.2.3. SNPs Associated with More than One Color Trait
3.2.4. SNPs Associated with Leaves
3.2.5. SNPs Associated with Pod Exterior Color
3.2.6. SNPs Associated with Pod Interior Color
3.2.7. Potential Candidate Genes Associated with SNPs
4. Discussion
4.1. Candidate Genes Associated with the Flavonoid Biosynthetic Pathway
4.2. Candidate Genes Associated with Photosynthetic Pathways
4.3. Candidate Genes from Other Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Commission Internationale de l’Éclairage. Colorimetry. CIE 15: Technical Report, 3rd ed.; CIE: Vienna, Austria, 2004. [Google Scholar]
- Patterson, C.A.; Maskus, H.; Dupasquier, C. Pulse Crops for Health. Cereal Foods World 2009, 54, 108–112. [Google Scholar] [CrossRef]
- Myers, J.R.; Baggett, J.R. Improvement of Snap Bean. In Common Bean Improvement in the Twenty-First Century; Singh, S.P., Ed.; Developments in Plant Breeding; Springer: Dordrecht, The Netherlands, 1999; pp. 289–329. [Google Scholar] [CrossRef]
- McClean, P.E.; Lee, R.K.; Otto, C.; Gepts, P.; Bassett, M.J. Molecular and phenotypic mapping of genes controlling seed coat pattern and color in common bean (Phaseolus vulgaris L.). J. Hered. 2002, 93, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Prakken, R. Inheritance of colour in Phaseolus vulgaris L. IV recombination within the “Complex Locus C.”. Meded Landbouwhogesch. Wagening. 1974, 24, 1–36. [Google Scholar]
- Bassett, M.J.; McClean, P.E. A brief review of the genetics of partly colored seed coats in common bean. Annu. Rep. Bean Improv. Coop. 2000, 43, 99–101. [Google Scholar]
- Beninger, C.W.; Hosfield, G.L.; Bassett, M.J. Flavonoid composition of three genotypes of dry bean (Phaseolus vulgaris) differing in seedcoat color. J. Am. Soc. Hortic. Sci. 1999, 124, 514–518. [Google Scholar] [CrossRef]
- Beninger, C.W.; Hosfield, G.L.; Bassett, M.J.; Owens, S. Chemical and morphological expression of the B and Asp seedcoat genes in Phaseolus vulgaris. J. Am. Soc. Hortic. Sci. 2000, 125, 52–58. [Google Scholar] [CrossRef]
- Prakken, R. Inheritance of colours and pod characters in Phaseolus vulgaris L. Genetica 1934, 16, 177–296. [Google Scholar] [CrossRef]
- Bassett, M.J. A new gene (Prpi-2) for intensified anthocyanin expression (IAE) syndrome in common bean and a reconciliation of gene symbols used by early investigators for purple pod and IAE syndrome. J. Am. Soc. Hortic. Sci. 2005, 130, 550–554. [Google Scholar] [CrossRef]
- Kyle, M.; Dickson, M. Linkage of hypersensitivity to five viruses with the B locus in Phaseolus vulgaris L. J. Hered. 1988, 79, 308–311. [Google Scholar] [CrossRef]
- Currence, T.M. A new pod color in snap beans. J. Hered. 1931, 22, 21–23. [Google Scholar] [CrossRef]
- Davis, J.; Myers, J.R.; McClean, P.; Lee, R. Staygreen (SGR), a candidate gene for the persistent color phenotype in common bean. Acta Hortic. 2010, 859, 99–102. [Google Scholar] [CrossRef]
- Hagerty, C.H.; Cuesta-Marcos, A.; Cregan, P.; Song, Q.; McClean, P.; Myers, J.R. Mapping snap bean pod and color traits, in a dry bean × snap bean recombinant inbred population. J. Am. Soc. Hortic. Sci. 2016, 141, 131–138. [Google Scholar] [CrossRef]
- Myers, J.; Davis, J.; Arkwazee, H.; Wallace, L.; Lee, R.; Moghaddam, S.; Mcclean, P. Why wax beans lack carotenoids. Annu. Rep. Bean Improv. Coop. 2019, 61, 29–30. [Google Scholar]
- Yang, X.; Liu, C.; Li, Y.; Yan, Z.; Liu, D.; Feng, G. Identification and fine genetic mapping of the golden pod gene (Pv-Ye) from the snap bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2021, 134, 3773–3784. [Google Scholar] [CrossRef]
- Myers, J.R.; Davis, J.W.; Yorgey, B.; Kean, D. Genetic analysis of processing traits in green bean (Phaseolus vulgaris). Acta Hortic. 2004, 637, 369–375. [Google Scholar] [CrossRef]
- García-Fernández, C.; Campa, A.; Ferreira, J.J. Dissecting the genetic control of seed coat color in a RIL population of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2021, 134, 3687–3698. [Google Scholar] [CrossRef]
- McClean, P.E.; Bett, K.E.; Stonehouse, R.; Lee, R.; Pflieger, S.; Moghaddam, S.M.; Geffroy, V.; Miklas, P.; Mamidi, S. White seed color in common bean (Phaseolus vulgaris) results from convergent evolution in the P (Pigment) gene. New Phytol. 2018, 219, 1112–1123. [Google Scholar] [CrossRef]
- Bassett, M.J.; Lee, R.; Symanietz, T.; McClean, P.E. Inheritance of reverse margo seedcoat pattern and allelism between the genes J for seedcoat color and L for partly colored seedcoat pattern in common bean. J. Am. Soc. Hortic. Sci. 2002, 127, 56–61. [Google Scholar] [CrossRef]
- Miklas, P. List of Genes—Phaseolus vulgaris L. Available online: http://www.bic.uprm.edu/wp-content/uploads/2019/10/Bean-Genes-List-2018-v2-1.pdf (accessed on 1 November 2018).
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Saballos, A.; Soler-Garzón, A.; Brooks, M.; Hart, J.P.; Lipka, A.E.; Miklas, P.; Peachey, R.E.; Tranel, P.J.; Williams, M.M. Multiple genomic regions govern tolerance to sulfentrazone in snap bean (Phaseolus vulgaris L.). Front. Agron. 2022, 4, 869770. [Google Scholar] [CrossRef]
- Hart, J.; Griffiths, P.D.; Mazourek, M.; Porch, T.; Gore, M.A.; Myers, J. Genomic Insight into the Breeding of Edible Podded Beans in a Snap Bean Association Panel (SnAP). In Proceedings of the Plant and Animal Genome Conference, San Diego, CA, USA, 10–14 January 2015. [Google Scholar]
- Celebioglu, B.; Myers, J.R.; Hart, J.P.; Porch, T.; Griffiths, P. Phenotypic variability for leaf and pod color within the Snap bean association panel. J. Am. Soc. Hortic. Sci. 2024, 149, 15–26. [Google Scholar] [CrossRef]
- Opassiri, R.; Pomthong, B.; Onkoksoong, T.; Akiyama, T.; Esen, A.; Ketudat Cairns, J.R. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-Glucosidase. BMC Plant Biol. 2006, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.-F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef]
- Jakoby, M.J.; Falkenhan, D.; Mader, M.T.; Brininstool, G.; Wischnitzki, E.; Platz, N.; Hudson, A.; Hülskamp, M.; Larkin, J.; Schnittger, A. Transcriptional profiling of mature arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008, 148, 1583–1602. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Abdulla, M.F.; Mostafa, K.; Seçgin, Z.; Yerlikaya, B.A.; Otur, Ç.; Gökdemir, G.; Kurt Kızıldoğan, A.; Al-Khayri, J.M.; Jain, S.M. Investigation and expression analysis of R2R3-MYBs and anthocyanin biosynthesis-related genes during seed color development of common bean (Phaseolus vulgaris). Plants 2022, 11, 3386. [Google Scholar] [CrossRef] [PubMed]
- Reinprecht, Y.; Yadegari, Z.; Perry, G.; Siddiqua, M.; Wright, L.; McClean, P.; Pauls, P. In silico comparison of genomic regions containing genes coding for enzymes and transcription factors for the phenylpropanoid pathway in Phaseolus vulgaris L. and Glycine max L. Merr. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Freixas Coutin, J.A.; Munholland, S.; Silva, A.; Subedi, S.; Lukens, L.; Crosby, W.L.; Pauls, K.P.; Bozzo, G.G. Proanthocyanidin accumulation and transcriptional responses in the seed coat of cranberry beans (Phaseolus vulgaris L.) with different susceptibility to postharvest darkening. BMC Plant Biol. 2017, 17, 89. [Google Scholar] [CrossRef]
- Erfatpour, M.; Navabi, A.; Pauls, K.P. Mapping the non-darkening trait from “Wit-Rood Boontje” in bean (Phaseolus vulgaris). Theor. Appl. Genet. 2018, 131, 1331–1344. [Google Scholar] [CrossRef]
- Erfatpour, M.; Pauls, K.P. A R2R3-MYB gene-based marker for the non-darkening seed coat trait in pinto and cranberry beans (Phaseolus vulgaris L.) derived from ‘Wit-Rood Boontje’. Theor. Appl. Genet. 2020, 133, 1977–1994. [Google Scholar] [CrossRef]
- Koinange, E.M.K.; Singh, S.P.; Gepts, P. Genetic control of the domestication syndrome in common bean. Crop. Sci. 1996, 36, 1037–1045. [Google Scholar] [CrossRef]
- García-Fernández, C.; Campa, A.; Garzón, A.S.; Miklas, P.; Ferreira, J.J. GWAS of pod morphological and color characters in common bean. BMC Plant Biol. 2021, 21, 184. [Google Scholar] [CrossRef]
- Tian, Y.; Zhong, R.; Wei, J.; Luo, H.; Eyal, Y.; Jin, H.; Wu, L.; Liang, K.; Li, Y.; Chen, S.; et al. Arabidopsis CHLOROPHYLLASE 1 protects young leaves from long-term photodamage by facilitating FtsH-mediated D1 degradation in photosystem II Repair. Mol. Plant 2021, 14, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.-M.; Chao, P.-Y.; Huang, M.-Y.; Yang, J.-H.; Yang, Z.-W.; Lin, K.-H.; Yang, C.-M. Chlorophyllase activity in green and non-green tissues of variegated plants. S. Afr. J. Bot. 2012, 81, 44–49. [Google Scholar] [CrossRef]
- Hu, B.; Zhu, J.; Wu, H.; Xu, K.; Zhai, H.; Guo, N.; Gao, Y.; Yang, J.; Zhu, D.; Xia, Z. Enhanced chlorophyll degradation triggers the pod degreening of “Golden Hook,” a special ecotype in common bean (Phaseolus vulgaris L.). Front. Genet. 2020, 11, 1074. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Yamada, T.; Masuda, Y.; Sato, Y.; Kobayashi, H.; Ueda, H.; Morita, R.; Nishimura, M.; Kitamura, K.; Kusaba, M. A green-cotyledon/stay-green mutant exemplifies the ancient whole-genome duplications in soybean. Plant Cell Physiol. 2014, 55, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Ibáñez, O.; Sharma, A.; Bykowski, M.; Borràs-Gas, G.; Behrendorff, J.B.Y.H.; Mellor, S.; Qvortrup, K.; Verdonk, J.C.; Bock, R.; Kowalewska, Ł.; et al. Curvature thylakoid 1 proteins modulate prolamellar body morphology and promote organized thylakoid biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2113934118. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Yamada, N.; Kagawa, T.; Yonekura, H.; Uyeda, T.Q.P.; Kadota, A.; Wada, M. Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 8860–8865. [Google Scholar] [CrossRef]
- Zhu, X.; Pan, Y.; Liu, Z.; Liu, Y.; Zhong, D.; Duan, Z.; Tian, Z.; Zhu, B.; Zhou, G. Mutation of YL results in a yellow leaf with chloroplast RNA editing defect in soybean. Int. J. Mol. Sci. 2020, 21, 4275. [Google Scholar] [CrossRef]
- Myers, J.R.; Wallace, L.T.; Mafi Moghaddam, S.; Kleintop, A.E.; Echeverria, D.; Thompson, H.J.; Brick, M.A.; Lee, R.; McClean, P.E. Improving the health benefits of snap bean: Genome-wide association studies of total phenolic content. Nutrients 2019, 11, 2509. [Google Scholar] [CrossRef]
- Martin, C.J. Characterization of Folate Biosynthesis Genes in Phaseolus vulgaris and Glycine max and Genome-Wide Association Study for Seed Folate Content in Phaseolus vulgaris. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2020. [Google Scholar]
- Kołton, A.; Długosz-Grochowska, O.; Wojciechowska, R.; Czaja, M. Biosynthesis regulation of folates and phenols in plants. Sci. Hortic. 2022, 291, 110561. [Google Scholar] [CrossRef]
- Weller, J.L.; Foo, E.M.; Hecht, V.; Ridge, S.; Vander Schoor, J.K.; Reid, J.B. Ethylene signaling influences light-regulated development in pea. Plant Physiol. 2015, 169, 115–124. [Google Scholar] [CrossRef]
- Izquierdo, P.; Astudillo, C.; Blair, M.W.; Iqbal, A.M.; Raatz, B.; Cichy, K.A. Meta-QTL analysis of seed iron and zinc concentration and content in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2018, 131, 1645–1659. [Google Scholar] [CrossRef] [PubMed]
- Jia, B. Genetic Analysis of Marsh Spot Resistance in Cranberry Common Bean. Master’s Thesis, University of Ottawa, Ottawa, ON, Canada, 2022. [Google Scholar]
- Zubimendi, J.P.; Martinatto, A.; Valacco, M.P.; Moreno, S.; Andreo, C.S.; Drincovich, M.F.; Tronconi, M.A. The complex allosteric and redox regulation of the fumarate hydratase and malate dehydratase reactions of Arabidopsis thaliana fumarase 1 and 2 gives clues for understanding the massive accumulation of fumarate. FEBS J. 2018, 285, 2205–2224. [Google Scholar] [CrossRef] [PubMed]
- Perez de Souza, L.; Scossa, F.; Proost, S.; Bitocchi, E.; Papa, R.; Tohge, T.; Fernie, A.R. Multi-tissue integration of transcriptomic and specialized metabolite profiling provides tools for assessing the common bean (Phaseolus vulgaris) metabolome. Plant J. 2019, 97, 1132–1153. [Google Scholar] [CrossRef] [PubMed]
- Erfatpour, M. Quantitative trait loci (QTL) and candidate gene identification for the non-darkening seed coat trait in dry beans (Phaseolus vulgaris L.). Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2021. [Google Scholar]
- Giordani, W. Dissecting the Genetic Architecture of Quantitative in Common Bean (Phaseolus vulgaris L.): Response to Root-Knot Nematode Infection and Seed Morphology. Ph.D. Thesis, University of São Paulo, São Paulo, Brazil, 2021. [Google Scholar]
- Vidak, M.; Lazarević, B.; Javornik, T.; Šatović, Z.; Carović-Stanko, K. Seed water absorption, germination, emergence and seedling phenotypic characterization of the common bean landraces differing in seed size and color. Seeds 2022, 1, 324–339. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Li, W.; Geng, J.; Li, S.; Yuan, X.; Gu, Y.; Zhong, J.; Zhang, Y.; Du, J. Genome-wide identification of the LBD transcription factor genes in common bean (Phaseolus vulgaris L.) and expression analysis under different abiotic stresses. J. Plant Interact. 2022, 17, 731–743. [Google Scholar] [CrossRef]
- Soltani, A.; Weraduwage, S.M.; Sharkey, T.D.; Lowry, D.B. Elevated temperatures cause loss of seed set in common bean (Phaseolus vulgaris L.) potentially through the disruption of source-sink relationships. BMC Genom. 2019, 20, 312. [Google Scholar] [CrossRef] [PubMed]
- Hayford, R.K.-A.; Ligaba-Osena, A.; Subramani, M.; Brown, A.; Melmaiee, K.; Hossain, K.; Kalavacharla, V. Characterization and expression analysis of common bean Histone Deacetylase 6 during development and cold stress response. Int. J. Genom. 2017, 2017, 2502691. [Google Scholar] [CrossRef]
- Toscano, S.; Trivellini, A.; Cocetta, G.; Bulgari, R.; Francini, A.; Romano, D.; Ferrante, A. Effect of preharvest abiotic stresses on the accumulation of bioactive compounds in horticultural produce. Front. Plant Sci. 2019, 10, 1212. [Google Scholar] [CrossRef]
- Liu, C.; Yang, X.; Yan, Z.; Liu, D.; Feng, G. Identification and characterization of a mutant Pv-Pur gene responsible for the purple phenotype of snap bean (Phaseolus vulgaris L.). Int. J. Mol. Sci. 2022, 23, 1265. [Google Scholar] [CrossRef]
- Duwadi, K.; Austin, R.S.; Mainali, H.R.; Bett, K.; Marsolais, F.; Dhaubhadel, S. Slow darkening of pinto bean seed coat is associated with significant metabolite and transcript differences related to proanthocyanidin biosynthesis. BMC Genom. 2018, 19, 260. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Iniguez, L.P.; Bucciarelli, B.; Roessler, J.; Schmutz, J.; McClean, P.E.; Jackson, S.A.; Hernandez, G.; Graham, M.A.; Stupar, R.M.; et al. A re-sequencing based assessment of genomic heterogeneity and fast neutron-induced deletions in a common bean cultivar. Front. Plant. Sci. 2013, 4, 210. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Li, L.; Li, Q.; Mao, H.; Wang, L.; Bie, Y.; Zeng, X.; Liao, L.; Wang, X.; Deng, H.; et al. Identification of photosynthesis characteristics and chlorophyll metabolism in leaves of citrus cultivar (Harumi) with varying degrees of chlorosis. Int. J. Mol. Sci. 2023, 24, 8394. [Google Scholar] [CrossRef] [PubMed]
- Wilker, J.L. Symbiotic Nitrogen Fixation in Heirloom, Landrace, and Modern Participatory-Bred and Conventionally-Bred Common Bean (Phaseolus vulgaris L.). Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2021. [Google Scholar]
- Giordani, W.; Gama, H.C.; Chiorato, A.F.; Garcia, A.A.F.; Vieira, M.L.C. Genome-wide association studies dissect the genetic architecture of seed shape and size in common bean. G3 2022, 12, jkac048. [Google Scholar] [CrossRef] [PubMed]
- Wahome, S.W.; Githiri, M.S.; Kinyanjui, P.K.; Toili, M.E.M.; Angenon, G. Genome-wide association study of variation in cooking time among common bean (Phaseolus vulgaris L.) accessions using diversity arrays technology markers. Legume Sci. 2023, e184. [Google Scholar] [CrossRef]
- Leitão, S.T.; Bicho, M.C.; Pereira, P.; Paulo, M.J.; Malosetti, M.; Araújo, S.d.S.; van Eeuwijk, F.; Vaz Patto, M.C. Common bean SNP alleles and candidate genes affecting photosynthesis under contrasting water regimes. Hortic. Res. 2021, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, A.; Zięba, P.; Gabryś, H. Sugar and light effects on the condition of the photosynthetic apparatus of Arabidopsis thaliana cultured in vitro. J. Plant. Growth Regul. 2012, 31, 90–101. [Google Scholar] [CrossRef]
- Gartner, W.; Bethke, P.C.; Kisha, T.J.; Nienhuis, J. Changes in sugar concentrations of seed and pod tissue during development in snap and dry beans (Phaseolus vulgaris L.). HortScience 2020, 55, 1692–1697. [Google Scholar] [CrossRef]
- Du, Y.; Li, W.; Geng, J.; Li, S.; Zhang, W.; Liu, X.; Hu, M.; Zhang, Z.; Fan, Y.; Yuan, X.; et al. Genome-wide identification of the SWEET gene family in Phaseolus vulgaris L. and their patterns of expression under abiotic stress. J. Plant Interact. 2022, 17, 390–403. [Google Scholar] [CrossRef]
- Freixas-Coutin, J.A. Transcriptomic and biochemical analyses of proanthocyanidin metabolism in seed coats of cranberry bean (Phaseolus vulgaris L.). Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2017. [Google Scholar]
- Foucher, J.; Ruh, M.; Préveaux, A.; Carrère, S.; Pelletier, S.; Briand, M.; Serre, R.-F.; Jacques, M.-A.; Chen, N.W.G. Common bean resistance to Xanthomonas is associated with upregulation of the salicylic acid pathway and downregulation of photosynthesis. BMC Genom. 2020, 21, 566. [Google Scholar] [CrossRef]
- Leitão, S.T.; Santos, C.; Araújo, S.d.S.; Rubiales, D.; Vaz Patto, M.C. Shared and tailored common bean transcriptomic responses to combined fusarium wilt and water deficit. Hortic. Res. 2021, 8, 149. [Google Scholar] [CrossRef] [PubMed]
- Rasool, S.; Mahajan, R.; Nazir, M.; Bhat, K.A.; Shikari, A.B.; Ali, G.; Bhat, B.; Bhat, B.A.; Shah, M.; Murtaza, I.; et al. SSR and GBS based GWAS study for identification of QTLs associated with nutritional elemental in common bean (Phaseolus vulgaris L.). Sci. Hortic. 2022, 306, 111470. [Google Scholar] [CrossRef]
- Baloch, F.S.; Nadeem, M.A.; Sönmez, F.; Habyarimana, E.; Mustafa, Z.; Karaköy, T.; Cömertpay, G.; Alsaleh, A.; Çiftçi, V.; Sun, S.; et al. Magnesium—A forgotten element: Phenotypic variation and genome wide association study in Turkish common bean germplasm. Front. Genet. 2022, 13, 848663. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; Tsai, S.M.; Chiorato, A.F.; da Silva Andrade, S.C.; Esteves, J.A.d.F.; Recchia, G.H.; Morais Carbonell, S.A. Analysis of the common bean (Phaseolus vulgaris L.) transcriptome regarding efficiency of phosphorus use. PLoS ONE 2019, 14, e0210428. [Google Scholar] [CrossRef] [PubMed]
- Ishida, J.K.; Caldas, D.G.G.; Oliveira, L.R.; Frederici, G.C.; Leite, L.M.P.; Mui, T.S. Genome-wide characterization of the NRAMP gene family in Phaseolus vulgaris provides insights into functional implications during common bean development. Genet. Mol. Biol. 2018, 41, 820–833. [Google Scholar] [CrossRef]
- Diaz, S.; Polania, J.; Ariza-Suarez, D.; Cajiao, C.; Grajales, M.; Raatz, B.; Beebe, S.E. Genetic correlation between Fe and Zn biofortification and yield components in a common bean (Phaseolus vulgaris L.). Front. Plant Sci. 2022, 12. [Google Scholar] [CrossRef]
| Gene | Chr | Function | Marker | Position (bp) | Source |
|---|---|---|---|---|---|
| pc | 2 | Encodes Mg dechelatase as the first step in the catabolism of chlorophyll | Phvul.002G153100 (v1.0 and v2.0) | 29,379,137–29,381,066 (v1.0) 30,648,161–30,650,090 (v2.0) | [13] |
| B | 2 | Controls production of anthocyanin precursors prior to dihydrokaempferol formation | ss715644998 | 48,634,623–48,634,743 | [14] |
| y | 2 | Pentatricopeptide repeat protein, or SUF family FE-S cluster assembly protein SufD | Phvul.002G004400, Phvul.002G006200 | [15,16] | |
| Ace | 6 | ? | [17] | ||
| V | 6 | Flavonoid 3′5′ hydroxylase (F3′5′H), a P450 enzyme required for the expression of dihydromyricetin-derived flavonoids | OD12800, Phvul.006G018800 | 9,288,398–9,288,419 | [4,18] |
| P | 7 | Basic helix-loop-helix protein regulating flavonoid pathway | Phvul.007G171333, Phvul.007G171466 | 28,752,132–28,774,743 | [19] |
| C | 8 | MYB113 transcription factor | OAP2700, Phvul.008G038400 | 9,694,328–9,695,007 | [4,18] |
| T | 9 | ? | OM19400 | 11,731,567–11,731,944 | [4] |
| j = L | 10 | Inhibits leuco-anthocyanidin production and reduces anthocyanin and flavonol production | OL4525 | 41,443,673–41,443,694 | [4,20] |
| Prp | ? | ? | [21] | ||
| arg | ? | ? | [21] |
| Leaf Position | Pod (Combined Years) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Middle | Upper | Interior | Exterior | ||||||
| Trait | And | Mid | And | Mid | And | Mid | And | Mid | And | Mid |
| L* | 28.5 | 30.9 | 30.5 | 31.5 | 29.6 | 28.4 | 80.5 | 80.8 | 78.3 | 77.4 |
| a* | 40.3 | 41.9 | 35.4 | 34.6 | 14.4 | 13.8 | 84.8 | 83.8 | 77.6 | 75.9 |
| b* | 33.4 | 35.4 | 32.9 | 33.0 | 25.6 | 24.8 | 58.1 | 55.9 | 75.0 | 72.5 |
| C* | 34.4 | 36.4 | 33.5 | 33.4 | 24.6 | 23.8 | 62.7 | 60.9 | 75.1 | 72.4 |
| H° | 31.3 | 34.2 | 29.3 | 30.1 | 26.1 | 25.9 | 83.8 | 83.0 | 74.5 | 72.4 |
| Chr | Position (bp) | −log10(p) | MAF | Traits | Effect | PVE (%) | H & B. P. Value |
|---|---|---|---|---|---|---|---|
| G19833 | |||||||
| 1 | 43,221,928 | 6.99 | 0.09 | a* (upper) | −0.61 | 33.64 | 1.94 × 10−3 |
| 43,965,518 | 10.98 | 0.05 | b* (middle) | 2.95 | 30.95 | 2.0 × 10−7 | |
| 49,802,297 | 6.70 | 0.09 | b* (lower) | −2.11 | 20.43 | 2.61 × 10−3 | |
| 49,802,297 | 6.13 | 0.09 | C* (lower) | −2.07 | 21.47 | 7.07 × 10−3 | |
| 2 | 33,626,429 | 6.25 | 0.07 | a* (middle) | 0.61 | 30.68 | 1.07 × 10−2 |
| 33,626,429 | 6.24 | 0.07 | C* (middle) | −1.92 | 13.90 | 1.11 × 10−2 | |
| 7 | 941,928 | 5.95 | 0.06 | L* (lower) | −1.40 | 12.12 | 1.08 × 10−2 |
| 1,336,909 | 6.17 | 0.07 | L* (upper) | −1.36 | 14.66 | 6.48 × 10−3 | |
| 9 | 22,912,954 | 7.24 | 0.05 | L* (upper) | 1.75 | 24.10 | 1.11 × 10−3 |
| 22,912,954 | 7.05 | 0.05 | H° (upper) | −1.65 | 56.85 | 1.70 × 10−3 | |
| 23,206,537 | 5.81 | 0.05 | L* (middle) | −1.42 | 54.93 | 2.94 × 10−2 | |
| 23,206,537 | 5.63 | 0.05 | C* (middle) | −1.86 | 40.35 | 2.29 × 10−2 | |
| 23,206,537 | 9.57 | 0.05 | b* (upper) | −3.08 | 55.54 | 5.20 × 10−6 | |
| 23,206,537 | 9.42 | 0.05 | C* (upper) | −3.03 | 54.82 | 7.32 × 10−6 | |
| 10 | 42,753,010 | 6.56 | 0.17 | b* (lower) | −1.38 | 11.02 | 2.61 × 10−3 |
| 42,753,010 | 6.49 | 0.17 | C* (lower) | −1.39 | 10.66 | 6.25 × 10−3 | |
| 5-593 | |||||||
| 1 | 46,046,838 | 5.95 | 0.05 | b* (lower) | 2.35 | 24.72 | 9.90 × 10−3 |
| 46,046,838 | 12.50 | 0.05 | b* (middle) | 3.35 | 31.63 | 8.41 × 10−9 | |
| 46,046,838 | 9.17 | 0.05 | L* (upper) | 2.17 | 53.42 | 1.79 × 10−5 | |
| 46,046,866 | 12.41 | 0.05 | C* (middle) | −3.37 | 31.65 | 1.02 × 10−8 | |
| 2 | 34,759,754 | 7.01 | 0.07 | a* (middle) | 0.64 | 28.88 | 2.62 × 10−3 |
| 3 | 2,905,494 | 8.07 | 0.05 | b* (middle) | 2.16 | 13.32 | 7.57 × 10−5 |
| 2,905,494 | 7.91 | 0.05 | C* (middle) | 2.16 | 13.10 | 1.09 × 10−4 | |
| 7 | 1,296,094 | 8.23 | 0.06 | L* (middle) | 1.53 | 16.53 | 1.56 × 10−4 |
| 9 | 25,192,705 | 7.82 | 0.05 | a* (upper) | 0.77 | 35.11 | 4.04 × 10−4 |
| 25,196,015 | 7.57 | 0.05 | H°(upper) | 1.59 | 57.00 | 7.10 × 10−4 | |
| 25,196,027 | 7.13 | 0.05 | L* (middle) | 1.58 | 17.40 | 6.58 × 10−4 | |
| 25,196,027 | 8.20 | 0.05 | H° (middle) | −1.82 | 22.67 | 8.28 × 10−5 | |
| 25,196,027 | 9.71 | 0.05 | b* (upper) | 2.97 | 33.56 | 5.19 × 10−6 | |
| 25,196,027 | 10.34 | 0.05 | C* (upper) | 3.23 | 55.11 | 1.21 × 10−6 | |
| Chr | Year | Position (bp) | −log10(p) | MAF | Traits | Effect | PVE (%) | H & B. P. Value |
|---|---|---|---|---|---|---|---|---|
| G19833 | ||||||||
| 1 | 2020 | 43,457,300 | 9.33 | 0.06 | L* | 4.36 | 21.68 | 4.46 × 10−6 |
| 2 | 807,178 | 14.48 | 0.23 | L* | 3.46 | 13.91 | 6.37 × 10−11 | |
| 2019 | 1,991,690 | 13.59 | 0.22 | a* | −1.20 | 26.94 | 2.45 × 10−10 | |
| 2020 | 1,991,690 | 16.40 | 0.22 | a* | −1.50 | 29.88 | 3.82 × 10−13 | |
| 2 years | 1,991,690 | 8.90 | 0.22 | a* | −0.96 | 17.53 | 6.07 × 10−6 | |
| 1,991,690 | 10.19 | 0.22 | C* | 0.78 | 26.40 | 3.05 × 10−7 | ||
| 2020 | 2,090,279 | 10.72 | 0.21 | a* | 4.40 | 18.57 | 6.08 × 10−8 | |
| 2019 | 2,090,300 | 9.78 | 0.21 | a* | −4.51 | 10.77 | 4.56 × 10−7 | |
| 2 years | 2,090,300 | 7.69 | 0.21 | C* | 3.67 | 21.22 | 4.34 × 10−5 | |
| 27,564,297 | 6.13 | 0.07 | L* | −6.29 | 24.21 | 1.76 × 10−3 | ||
| 2019 | 27,727,002 | 15.10 | 0.08 | L* | 4.55 | 11.12 | 7.60 × 10−12 | |
| 2 years | 27,727,002 | 15.67 | 0.08 | L* | 11.06 | 25.78 | 2.05 × 10−12 | |
| 2019 | 30,590,620 | 21.91 | 0.06 | b* | 5.23 | 10.75 | 2.34 × 10−18 | |
| 3 | 2020 | 10,297,623 | 8.63 | 0.14 | b* | 6.88 | 45.21 | 1.49 × 10−5 |
| 10,888,291 | 10.97 | 0.13 | b* | −1.29 | 28.25 | 1.03 × 10−7 | ||
| 42,856,778 | 12.15 | 0.07 | H° | −56.50 | 27.91 | 1.23 × 10−9 | ||
| 43,092,849 | 15.52 | 0.06 | H° | 10.11 | 32.04 | 2.89 × 10−12 | ||
| 4 | 2020 | 152,406 | 8.72 | 0.05 | C* | 2.06 | 12.11 | 1.22 × 10−5 |
| 8 | 2 years | 706,796 | 11.61 | 0.36 | H° | −31.60 | 10.89 | 7.76 × 10−9 |
| 706,882 | 15.99 | 0.35 | H° | −6.82 | 22.47 | 6.70 × 10−13 | ||
| 1,732,687 | 6.95 | 0.12 | a* | −1.66 | 24.36 | 3.09 × 10−4 | ||
| 1,752,346 | 5.64 | 0.24 | a* | 5.95 | 16.24 | 2.93 × 10−3 | ||
| 9 | 2020 | 10,765,350 | 6.04 | 0.07 | C* | −1.99 | 11.92 | 2.58 × 10−3 |
| 10 | 2 years | 4,715,815 | 9.58 | 0.36 | b* | −0.82 | 10.30 | 1.70 × 10−6 |
| 11 | 2019 | 51,718,220 | 9.34 | 0.06 | b* | 4.20 | 11.76 | 1.09 × 10−6 |
| 5-593 | ||||||||
| 1 | 2020 | 46,993,079 | 13.37 | 0.05 | L* | 5.43 | 14.31 | 3.75 × 10−10 |
| 50,939,307 | 27.26 | 0.08 | H° | 75.89 | 29.33 | 1.47 × 10−23 | ||
| 51,026,803 | 26.04 | 0.07 | H° | −73.80 | 28.52 | 1.21 × 10−22 | ||
| 2 years | 51,026,803 | 8.62 | 0.07 | C* | 1.77 | 19.89 | 2.27 × 10−6 | |
| 2 | 2019 | 1,604,407 | 15.73 | 0.23 | L* | 3.08 | 10.18 | 4.93 × 10−12 |
| 2020 | 1,604,407 | 18.75 | 0.23 | L* | 4.05 | 12.19 | 2.35 × 10−15 | |
| 2019 | 23,372,981 | 8.08 | 0.10 | a* | −3.07 | 13.50 | 2.21 × 10−5 | |
| 23,874,856 | 20.54 | 0.08 | H° | 21.39 | 16.29 | 2.55 × 10−17 | ||
| 2020 | 23,874,856 | 8.78 | 0.08 | b* | −1.42 | 15.70 | 8.81 × 10−6 | |
| 2019 | 23,874,857 | 8.24 | 0.08 | a* | −3.36 | 15.43 | 1.69 × 10−5 | |
| 2020 | 23,874,857 | 7.99 | 0.08 | a* | −3.11 | 36.69 | 8.25 × 10−4 | |
| 2 years | 23,874,857 | 7.89 | 0.08 | a* | −3.17 | 15.12 | 6.88 × 10−5 | |
| 2020 | 25,183,895 | 6.81 | 0.10 | a* | 3.21 | 14.55 | 8.40 × 10−4 | |
| 29,034,302 | 12.47 | 0.06 | H° | 99.32 | 11.30 | 2.56 × 10−10 | ||
| 2019 | 31,662,002 | 7.15 | 0.05 | L* | 4.17 | 13.70 | 3.58 × 10−4 | |
| 2020 | 31,662,002 | 19.09 | 0.05 | L* | 6.21 | 19.57 | 2.17 × 10−15 | |
| 2 years | 31,662,002 | 17.07 | 0.05 | L* | 5.59 | 16.50 | 1.13 × 10−13 | |
| 31,662,002 | 24.98 | 0.05 | H° | −58.80 | 23.82 | 9.17 × 10−22 | ||
| 31,744,956 | 18.80 | 0.05 | H° | −44.37 | 11.30 | 1.04 × 10−15 | ||
| 3 | 2019 | 36,729,340 | 30.11 | 0.08 | b* | 8.36 | 29.56 | 2.04 × 10−26 |
| 36,729,340 | 45.77 | 0.08 | H° | −47.89 | 23.78 | 4.50 × 10−42 | ||
| 2020 | 36,729,340 | 10.75 | 0.08 | b* | 5.82 | 16.37 | 1.56 × 10−7 | |
| 36,729,340 | 21.31 | 0.08 | C* | 7.12 | 24.58 | 6.44 × 10−18 | ||
| 2 years | 36,729,340 | 24.60 | 0.08 | b* | 7.78 | 42.38 | 6.72 × 10−21 | |
| 36,729,340 | 57.34 | 0.08 | H° | −46.96 | 22.01 | 1.22 × 10−53 | ||
| 2019 | 36,757,348 | 28.35 | 0.10 | b* | −7.39 | 20.16 | 5.97 × 10−25 | |
| 36,757,348 | 38.52 | 0.10 | H° | 39.62 | 20.75 | 4.04 × 10−35 | ||
| 2020 | 36,757,348 | 10.57 | 0.10 | b* | −4.87 | 17.79 | 1.77 × 10−7 | |
| 36,757,348 | 23.78 | 0.10 | C* | −6.96 | 13.82 | 4.41 × 10−20 | ||
| 2 years | 36,757,348 | 24.23 | 0.10 | b* | −7.39 | 28.44 | 7.79 × 10−21 | |
| 36,757,348 | 43.30 | 0.10 | H° | 36.47 | 13.64 | 6.63 × 10−40 | ||
| 4 | 2019 | 51,719,381 | 9.86 | 0.07 | a* | 4.04 | 14.47 | 1.23 × 10−6 |
| 2 years | 51,719,381 | 6.78 | 0.07 | a* | 3.18 | 19.24 | 6.28 × 10−4 | |
| Chr | Year | Position (bp) | −log10(p) | MAF | Traits | Effect | PVE (%) | H & B. P. Value |
|---|---|---|---|---|---|---|---|---|
| G19833 | ||||||||
| 1 | 2019 | 47,592,371 | 7.21 | 0.07 | C* | 2.00 | 13.9 | 3.96 × 10−4 |
| 2 years | 48,364,135 | 7.14 | 0.06 | C* | 1.64 | 15.2 | 4.62 × 10−4 | |
| 2 | 2019 | 790,825 | 30.81 | 0.23 | a* | −2.73 | 19.9 | 2.95 × 10−27 |
| 2020 | 790,825 | 39.05 | 0.23 | H° | 4.28 | 18.1 | 1.69 × 10−35 | |
| 2 years | 790,825 | 24.53 | 0.23 | L* | −3.71 | 19.1 | 5.66 × 10−21 | |
| 2020 | 807,178 | 23.44 | 0.23 | L* | 3.94 | 14.0 | 6.88 × 10−20 | |
| 807,178 | 39.61 | 0.23 | a* | 3.10 | 16.1 | 4.70 × 10−36 | ||
| 2 years | 807,178 | 38.40 | 0.23 | a* | 2.94 | 15.0 | 7.67 × 10−35 | |
| 807,178 | 43.70 | 0.23 | H° | −4.22 | 13.9 | 3.79 × 10−40 | ||
| 2019 | 2,190,426 | 10.30 | 0.07 | a* | 2.01 | 11.7 | 4.85 × 10−7 | |
| 2 years | 2,190,426 | 12.76 | 0.07 | L* | 3.29 | 15.9 | 1.68 × 10−9 | |
| 5 | 2019 | 11,368,205 | 7.62 | 0.22 | H° | 0.99 | 19.0 | 1.23 × 10−4 |
| 6 | 2 years | 14,661,470 | 7.87 | 0.44 | b* | −0.82 | 23.8 | 2.59 × 10−4 |
| 2020 | 14,665,606 | 9.89 | 0.44 | C* | 1.19 | 12.9 | 2.48 × 10−6 | |
| 18,684,799 | 5.71 | 0.19 | C* | 0.83 | 16.3 | 1.86 × 10−2 | ||
| 5-593 | ||||||||
| 2 | 2 years | 1,587,342 | 16.18 | 0.23 | C* | 1.40 | 21.4 | 1.75 × 10−12 |
| 2019 | 1,604,407 | 36.65 | 0.23 | a* | 2.85 | 15.8 | 5.96 × 10−33 | |
| 1,604,407 | 37.08 | 0.23 | H° | −4.38 | 22.8 | 2.23 × 10−33 | ||
| 2020 | 1,604,407 | 30.29 | 0.23 | L* | 4.54 | 13.3 | 1.37 × 10−26 | |
| 1,604,407 | 42.80 | 0.23 | a* | 3.28 | 17.8 | 4.19 × 10−39 | ||
| 1,604,407 | 45.69 | 0.23 | H° | −4.79 | 20.2 | 5.38 × 10−42 | ||
| 2 years | 1,604,407 | 23.48 | 0.23 | L* | 3.77 | 20.9 | 8.70 × 10−20 | |
| 1,604,407 | 41.27 | 0.23 | a* | 3.04 | 17.6 | 1.41 × 10−37 | ||
| 1,604,407 | 46.11 | 0.23 | H° | −4.65 | 18.0 | 2.06 × 10−42 | ||
| 2020 | 2,979,708 | 14.51 | 0.07 | H° | −3.23 | 10.6 | 4.07 × 10−11 | |
| Chr | Allele | Traits | Position (bp) | −log10(p) | MAF | Effect | PVE (%) | H & B. P. Value | Candidate Genes | Best.Hit |
|---|---|---|---|---|---|---|---|---|---|---|
| G19833 | ||||||||||
| 2 | C/T | L*.2 year | 27,727,002 | 15.67 | 0.08 | 11.06 | 25.8 | 2.05 × 10−12 | Phvul.002G132400 | AT1G32100 |
| Phvul.002G132800 | AT1G32080 | |||||||||
| 3 | G/A | H°.2020 | 42,856,778 | 12.15 | 0.07 | −56.50 | 27.9 | 1.23 × 10−9 | Phvul.003G202200 | AT5G63050 |
| Phvul.003G202400 | AT1G17100 | |||||||||
| Phvul.003G203400 | ||||||||||
| G/C | H°.2020 | 43,092,849 | 15.52 | 0.06 | 10.11 | 32.0 | 2.89 × 10−12 | Phvul.003G203700 | AT5G63100 | |
| Phvul.003G203800 | AT5G63110 | |||||||||
| Phvul.003G203900 | AT5G26660 | |||||||||
| Phvul.003G204700 | AT3G11330 | |||||||||
| Phvul.003G205000 | AT4G24690 | |||||||||
| Phvul.003G205300 | AT3G48250 | |||||||||
| Phvul.003G205400 | AT3G48250 | |||||||||
| Phvul.003G205800 | ||||||||||
| Phvul.003G206100 | AT3G48270 | |||||||||
| 8 | G/A | H°.2 year | 706,796 | 11.61 | 0.36 | −31.60 | 10.9 | 7.76 × 10−9 | Phvul.008G006600 | AT5G65560 |
| Phvul.008G006700 | AT5G38720 | |||||||||
| Phvul.008G006800 | AT2G19540 | |||||||||
| Phvul.008G007500 | AT5G16150 | |||||||||
| Phvul.008G007600 | AT2G39060 | |||||||||
| Phvul.008G007700 | AT5G16180 | |||||||||
| Phvul.008G008400 | AT1G66840 | |||||||||
| 5-593 | ||||||||||
| 1 | G/T | H°.2020 | 50,939,307 | 27.26 | 0.08 | 75.89 | 29.3 | 1.47 × 10−23 | Pv5-593.01G220900 | AT5G26600 |
| Pv5-593.01G221300 | AT1G56720 | |||||||||
| Pv5-593.01G221400 | ||||||||||
| Pv5-593.01G221500 | AT1G09430 | |||||||||
| Pv5-593.01G221600 | AT3G07100 | |||||||||
| Pv5-593.01G221800 | AT1G56700 | |||||||||
| Pv5-593.01G221900 | AT1G09420 | |||||||||
| Pv5-593.01G222100 | AT1G09390 | |||||||||
| Pv5-593.01G222400 | AT1G09340 | |||||||||
| Pv5-593.01G222600 | AT1G56590 | |||||||||
| Pv5-593.01G222700 | AT4G15510 | |||||||||
| A/G | H°.2020 | 51,026,803 | 26.04 | 0.07 | −73.80 | 28.5 | 1.21 × 10−22 | Pv5-593.01G221400 | ||
| Pv5-593.01G221500 | AT1G09430 | |||||||||
| Pv5-593.01G221600 | AT3G07100 | |||||||||
| Pv5-593.01G221800 | AT1G56700 | |||||||||
| Pv5-593.01G221900 | AT1G09420 | |||||||||
| Pv5-593.01G222100 | AT1G09390 | |||||||||
| Pv5-593.01G222400 | AT1G09340 | |||||||||
| Pv5-593.01G222600 | AT1G56590 | |||||||||
| Pv5-593.01G222700 | AT4G15510 | |||||||||
| Pv5-593.01G223300 | AT1G56500 | |||||||||
| 2 | T/G | H°.2019 | 23,874,856 | 20.54 | 0.08 | 21.39 | 16.3 | 2.55 × 10−17 | Pv5-593.02G108100 | AT5G66280 |
| Pv5-593.02G108300 | AT1G68920 | |||||||||
| A/T | H°.2020 | 29,034,302 | 12.47 | 0.06 | 99.32 | 11.3 | 2.56 × 10−10 | Pv5-593.02G134200 | AT4G38100 | |
| Pv5-593.02G134400 | AT2G22500 | |||||||||
| Pv5-593.02G134700 | AT2G22530 | |||||||||
| Pv5-593.02G134900 | AT5G10470 | |||||||||
| G/C | H°.2 year | 31,662,002 | 24.98 | 0.05 | −58.80 | 23.8 | 9.17 × 10−22 | Pv5-593.02G153100 | AT1G63430 | |
| Pv5-593.02G153300 | AT1G63440 | |||||||||
| Pv5-593.02G153400 | ||||||||||
| Pv5-593.02G154000 | AT1G63490 | |||||||||
| Pv5-593.02G154100 | ||||||||||
| G/T | H°.2 year | 31,744,956 | 18.80 | 0.05 | −44.37 | 11.3 | 1.04 × 10−15 | Pv5-593.02G153100 | AT1G63430 | |
| Pv5-593.02G153300 | AT1G63440 | |||||||||
| Pv5-593.02G153400 | ||||||||||
| Pv5-593.02G154000 | AT1G63490 | |||||||||
| Pv5-593.02G154100 | ||||||||||
| Pv5-593.02G155000 | AT5G41380 | |||||||||
| 3 | T/C | H°.2019 | 36,729,340 | 45.77 | 0.08 | −47.89 | 23.8 | 4.50 × 10−42 | Pv5-593.03G141800 | AT2G18850 |
| T/C | H°.2 year | 36,729,340 | 57.34 | 0.08 | −46.96 | 22.0 | 1.22 × 10−53 | Pv5-593.03G142100 | AT5G57490 | |
| T/C | H°.2019 | 36,757,348 | 38.52 | 0.10 | 39.62 | 20.8 | 4.04 × 10−35 | Pv5-593.03G142300 | AT5G57510 | |
| T/C | H°.2 year | 36,757,348 | 43.30 | 0.10 | 36.47 | 13.6 | 6.63 × 10−40 | Pv5-593.03G142400 | AT1G51680 | |
| Pv5-593.03G142500 | ||||||||||
| Pv5-593.03G142800 | ||||||||||
| Pv5-593.03G142900 | AT5G57560 | |||||||||
| Pv5-593.03G143000 | AT5G57560 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celebioglu, B.; Hart, J.P.; Porch, T.; Griffiths, P.; Myers, J.R. Genome-Wide Association Study to Identify Possible Candidate Genes of Snap Bean Leaf and Pod Color. Genes 2023, 14, 2234. https://doi.org/10.3390/genes14122234
Celebioglu B, Hart JP, Porch T, Griffiths P, Myers JR. Genome-Wide Association Study to Identify Possible Candidate Genes of Snap Bean Leaf and Pod Color. Genes. 2023; 14(12):2234. https://doi.org/10.3390/genes14122234
Chicago/Turabian StyleCelebioglu, Burcu, John P. Hart, Timothy Porch, Phillip Griffiths, and James R. Myers. 2023. "Genome-Wide Association Study to Identify Possible Candidate Genes of Snap Bean Leaf and Pod Color" Genes 14, no. 12: 2234. https://doi.org/10.3390/genes14122234
APA StyleCelebioglu, B., Hart, J. P., Porch, T., Griffiths, P., & Myers, J. R. (2023). Genome-Wide Association Study to Identify Possible Candidate Genes of Snap Bean Leaf and Pod Color. Genes, 14(12), 2234. https://doi.org/10.3390/genes14122234






