Lambda CI Binding to Related Phage Operator Sequences Validates Alignment Algorithm and Highlights the Importance of Overlooked Bonds
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sequence Analysis
2.3. Protein Purification
2.4. Electrophoretic Mobility Shift Assays (EMSAs)
2.5. Data Analysis
2.6. Pattern Alignment
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farrokh, C.; Jordan, K.; Auvray, F.; Glass, K.; Oppegaard, H.; Raynaud, S.; Thevenot, D.; Condron, R.; De Reu, K.; Govaris, A.; et al. Review of Shiga-Toxin-Producing Escherichia coli (STEC) and Their Significance in Dairy Production. Int. J. Food Microbiol. 2013, 162, 190–212. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.L.; Leibowitz, C.S.; Kurosawa, S.; Stearns-Kurosawa, D.J. Shiga Toxins and the Pathophysiology of Hemolytic Uremic Syndrome in Humans and Animals. Toxins 2012, 4, 1261–1287. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lewis, D.E.A.; Adhya, S. The Developmental Switch in Bacteriophage λ: A Critical Role of the Cro Protein. J. Mol. Biol. 2018, 430, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Sedhom, J.; Kinser, J.; Solomon, L.A. Alignment of Major-Groove Hydrogen Bond Arrays Uncovers Shared Information between Different DNA Sequences That Bind the Same Protein. NAR Genom. Bioinform. 2022, 4, lqac101. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Rooks, D.J.; Fogg, P.C.; Darby, A.C.; Thomson, N.R.; McCarthy, A.J.; Allison, H.E. Comparative Genomics of Shiga Toxin Encoding Bacteriophages. BMC Genom. 2012, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Nejman-Faleńczyk, B.; Bloch, S.; Węgrzyn, A.; Węgrzyn, G.; Donaldson, L.W. Ea22 Proteins from Lambda and Shiga Toxin-Producing Bacteriophages Balance Structural Diversity with Functional Similarity. ACS Omega 2020, 5, 12236–12244. [Google Scholar] [CrossRef] [PubMed]
- Sarkar-Banerjee, S.; Goyal, S.; Gao, N.; Mack, J.; Thompson, B.; Dunlap, D.; Chattopadhyay, K.; Finzi, L. Specifically Bound Lambda Repressor Dimers Promote Adjacent Non-Specific Binding. PLoS ONE 2018, 13, e0194930. [Google Scholar] [CrossRef]
- Ptashne, M. The Chemistry of Regulation of Genes and Other Things. J. Biol. Chem. 2014, 289, 5417–5435. [Google Scholar] [CrossRef]
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2004; ISBN 978-0-87969-717-4. [Google Scholar]
- Stayrook, S.; Jaru-Ampornpan, P.; Ni, J.; Hochschild, A.; Lewis, M. Crystal Structure of the λ Repressor and a Model for Pairwise Cooperative Operator Binding. Nature 2008, 452, 1022–1025. [Google Scholar] [CrossRef]
- Fattah, K.R.; Mizutani, S.; Fattah, F.J.; Matsushiro, A.; Sugino, Y. A Comparative Study of the Immunity Region of Lambdoid Phages Including Shiga-Toxin-Converting Phages: Molecular Basis for Cross Immunity. Genes Genet. Syst. 2000, 75, 223–232. [Google Scholar] [CrossRef]
- Benson, N.; Adams, C.; Youderian, P. Mutant Lambda Repressors with Increased Operator Affinities Reveal New, Specific Protein-DNA Contacts. Genetics 1992, 130, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.; Le, P.; Zurla, C.; Finzi, L.; Adhya, S. Multilevel Autoregulation of λ Repressor Protein CI by DNA Looping In Vitro. Proc. Natl. Acad. Sci. USA 2011, 108, 14807–14812. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Guo, J. New Insights into Protein–DNA Binding Specificity from Hydrogen Bond Based Comparative Study. Nucleic Acids Res. 2019, 47, 11103–11113. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.; Thukral, S.; Krishnan, S.; Chakrobarty, M.; Gupta, S.; Manghani, C.; Rani, V. DNA–Protein Interactions: Methods for Detection and Analysis. Mol. Cell. Biochem. 2012, 365, 279–299. [Google Scholar] [CrossRef]
- Solomon, L.A.; Witten, J.; Kodali, G.; Moser, C.C.; Dutton, P.L. Tailorable Tetrahelical Bundles as a Toolkit for Redox Studies. J. Phys. Chem. B 2022, 126, 8177–8187. [Google Scholar] [CrossRef]
- Gao, N.; Shearwin, K.; Mack, J.; Finzi, L.; Dunlap, D. Purification of Bacteriophage Lambda Repressor. Protein Expr. Purif. 2013, 91, 30–36. [Google Scholar] [CrossRef][Green Version]
- Heffler, M.A.; Walters, R.D.; Kugel, J.F. Using Electrophoretic Mobility Shift Assays to Measure Equilibrium Dissociation Constants: GAL4-P53 Binding DNA as a Model System. Biochem. Mol. Biol. Educ. 2012, 40, 383–387. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Ream, J.A.; Lewis, L.K.; Lewis, K.A. Rapid Agarose Gel Electrophoretic Mobility Shift Assay for Quantitating Protein: RNA Interactions. Anal. Biochem. 2016, 511, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 47, D94–D99. [Google Scholar] [CrossRef] [PubMed]
- Glazko, G.; Makarenkov, V.; Liu, J.; Mushegian, A. Evolutionary History of Bacteriophages with Double-Stranded DNA Genomes. Biol. Direct 2007, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Astromoff, A.; Ptashne, M. A Variant of Lambda Repressor with an Altered Pattern of Cooperative Binding to DNA Sites. Proc. Natl. Acad. Sci. USA 1995, 92, 8110–8114. [Google Scholar] [CrossRef]
- Bell, C.E.; Frescura, P.; Hochschild, A.; Lewis, M. Crystal Structure of the λ Repressor C-terminal Domain Provides a Model for Cooperative Operator Binding. Cell 2000, 101, 801–811. [Google Scholar] [CrossRef]
- Hochschild, A.; Douhan, J.; Ptashne, M. How λ Repressor and λ Cro Distinguish between OR1 and OR3. Cell 1986, 47, 807–816. [Google Scholar] [CrossRef]
- Hochschild, A.; Lewis, M. The Bacteriophage λ CI Protein Finds an Asymmetric Solution. Curr. Opin. Struct. Biol. 2009, 19, 79–86. [Google Scholar] [CrossRef]
- Siggers, T.; Gordân, R. Protein–DNA Binding: Complexities and Multi-Protein Codes. Nucleic Acids Res. 2014, 42, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Emamjomeh, A.; Choobineh, D.; Hajieghrari, B.; MahdiNezhad, N.; Khodavirdipour, A. DNA–Protein Interaction: Identification, Prediction and Data Analysis. Mol. Biol. Rep. 2019, 46, 3571–3596. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M. Amino Acid-Base Interactions: A Three-Dimensional Analysis of Protein-DNA Interactions at an Atomic Level. Nucleic Acids Res. 2001, 29, 2860–2874. [Google Scholar] [CrossRef] [PubMed]
- Garvie, C.W.; Wolberger, C. Recognition of Specific DNA Sequences. Mol. Cell 2001, 8, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Meyer, B.J.; Ptashne, M. Interactions between DNA-Bound Repressors Govern Regulation by the λ Phage Repressor. Proc. Natl. Acad. Sci. USA 1979, 76, 5061–5065. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein–Nucleic Acid Interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Buratowski, S.; Chodosh, L.A. Mobility Shift DNA -Binding Assay Using Gel Electrophoresis. CP Mol. Biol. 1996, 36. [Google Scholar] [CrossRef]
- Beamer, L.J.; Pabo, C.O. Refined 1.8 Å Crystal Structure of the λ Repressor-Operator Complex. J. Mol. Biol. 1992, 227, 177–196. [Google Scholar] [CrossRef]
- Kalodimos, C.G.; Biris, N.; Bonvin, A.M.J.J.; Levandoski, M.M.; Guennuegues, M.; Boelens, R.; Kaptein, R. Structure and Flexibility Adaptation in Nonspecific and Specific Protein-DNA Complexes. Science 2004, 305, 386–389. [Google Scholar] [CrossRef]
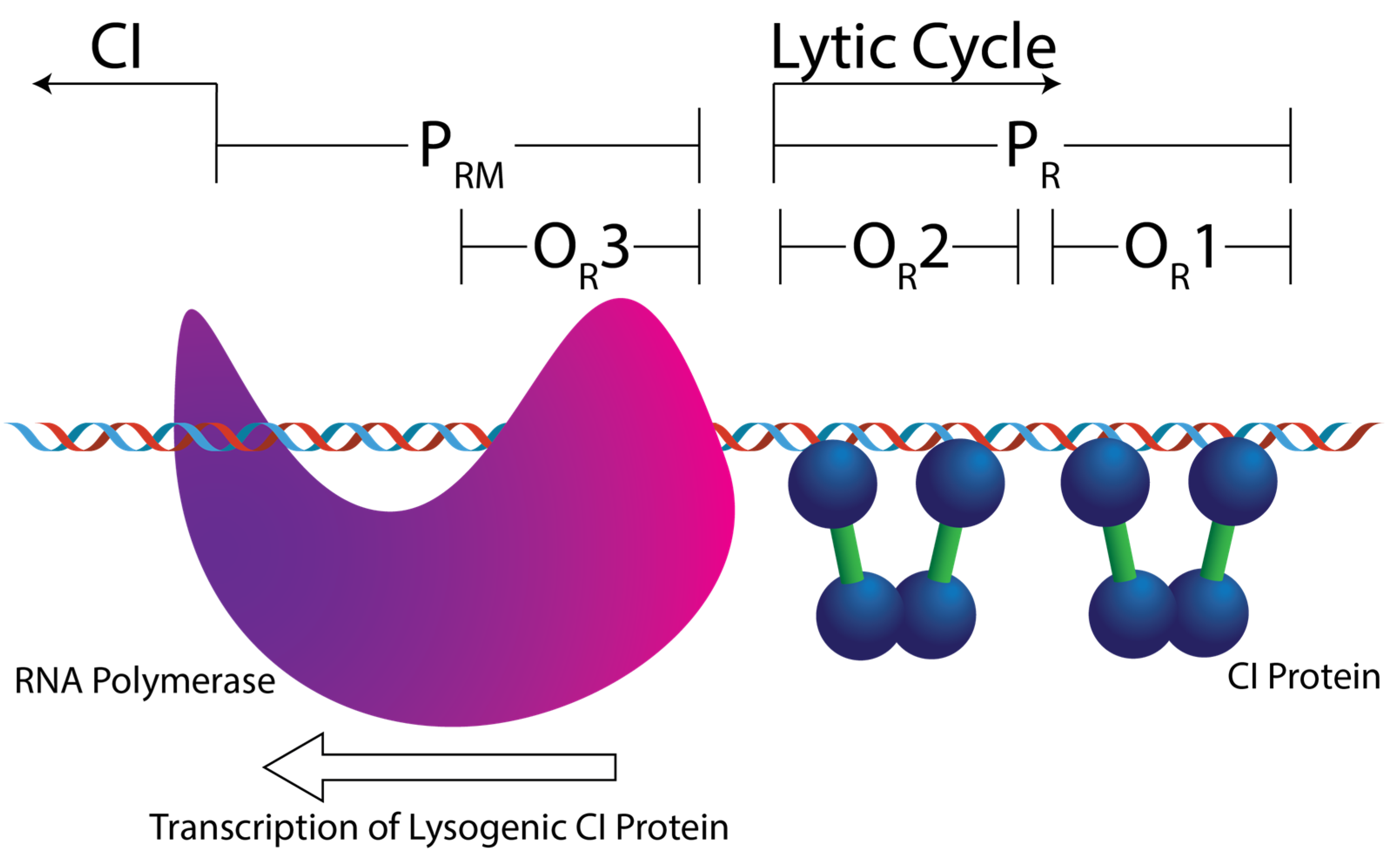

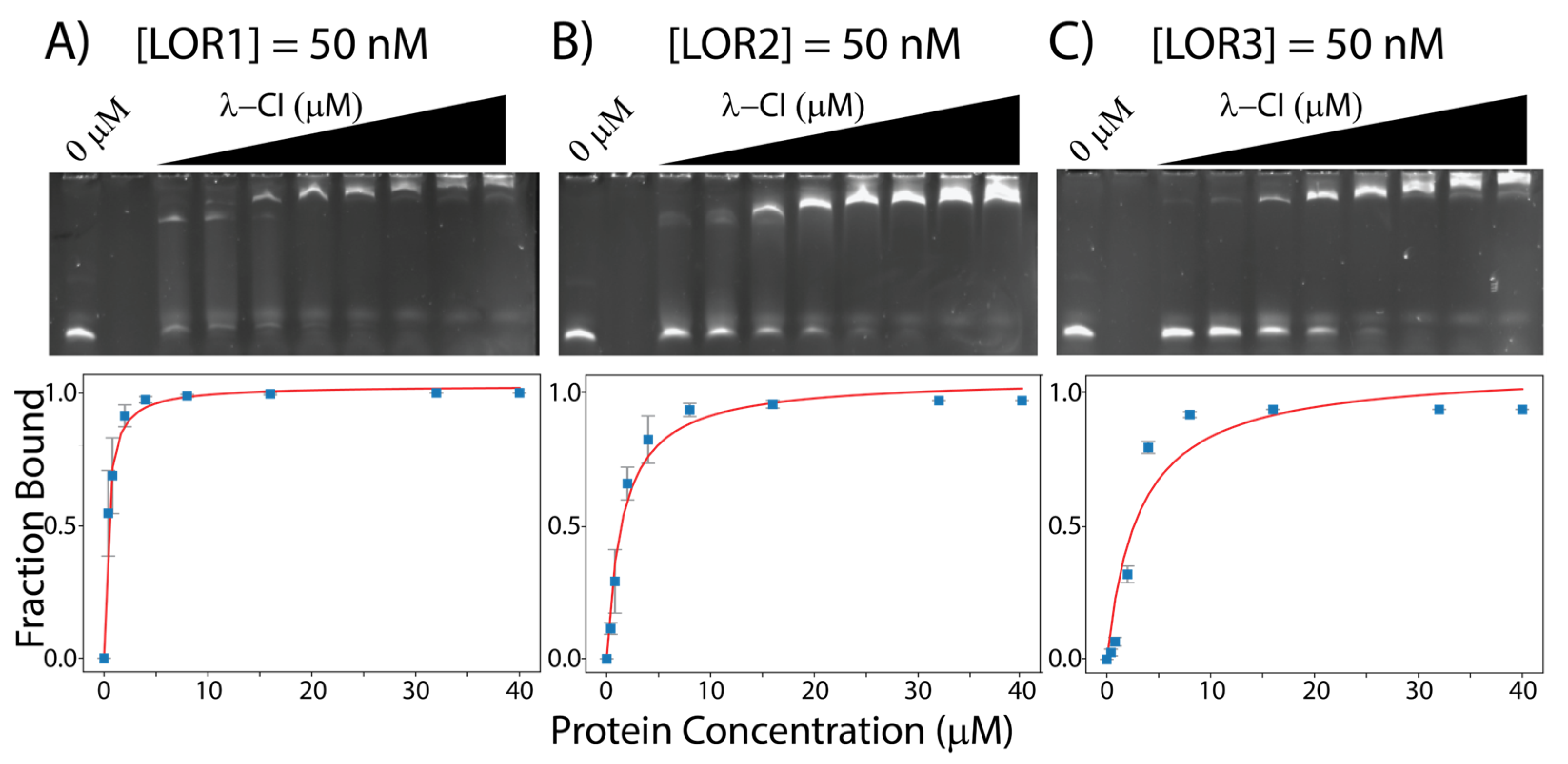
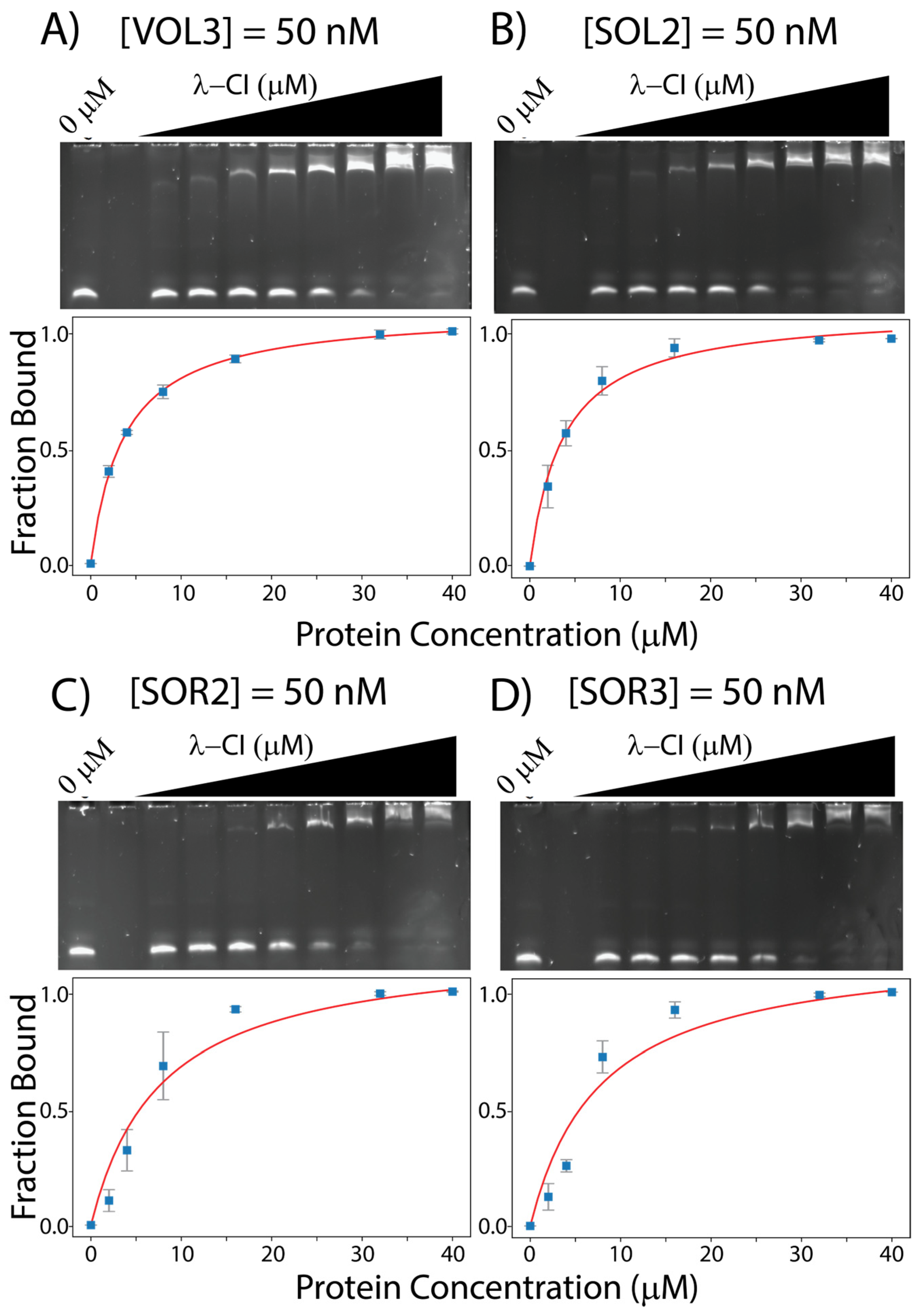

| Operator Site | OR1 | OR2 | OR3 | OL1 | OL2 | OL3 |
|---|---|---|---|---|---|---|
| λ-Phage | 0.35 ± 0.03 | 1.55 ± 0.29 | 3.07 ± 1.13 | 0.21 ± 0.02 | 1.50 ± 0.17 | 1.21 ± 0.08 |
| VT2-SA | 5.15 ± 2.39 | 6.39 ± 2.75 | 4.41 ± 0.99 | 5.37 ± 1.16 | 4.02 ± 0.16 | 3.67 ± 0.10 |
| Stx2I | 7.27 ± 2.39 | 7.67 ± 2.62 | 7.68 ± 3.09 | 7.11 ± 1.27 | 3.68 ± 0.47 | 4.26 ± 0.27 |
| KDapp (µM) | Consensus %Match | Distinct %Match | Specific Bonds | |
|---|---|---|---|---|
| VOL3 | 3.67 ± 0.10 | 73.9% | 64.3% | 6D, 12A |
| SOL2 | 3.68 ± 0.47 | 43.5% | 50.0% | 6D |
| VOL2 | 4.02 ± 0.16 | 78.3% | 64.3% | 6D, 12A |
| SOL3 | 4.26 ± 0.27 | 52.2% | 57.1% | 6D, 7D, 12A |
| VOR3 | 4.41 ± 0.99 | 69.6% | 64.3% | 6D, 12A |
| VOR1 | 5.15 ± 2.39 | 73.9% | 78.6% | 6D, 12A |
| VOL1 | 5.37 ± 1.16 | 73.9% | 78.6% | 6D, 12A |
| VOR2 | 6.39 ± 2.75 | 65.2% | 57.1% | 6D, 12A |
| SOL1 | 7.11 ± 1.27 | 69.6% | 64.3% | 7D |
| SOR1 | 7.27 ± 2.39 | 39.1% | 42.9% | 7D |
| SOR2 | 7.67 ± 2.62 | 26.1% | 28.6% | 6D, 12A |
| SOR3 | 7.68 ± 3.09 | 65.2% | 64.3% | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedhom, J.; Solomon, L.A. Lambda CI Binding to Related Phage Operator Sequences Validates Alignment Algorithm and Highlights the Importance of Overlooked Bonds. Genes 2023, 14, 2221. https://doi.org/10.3390/genes14122221
Sedhom J, Solomon LA. Lambda CI Binding to Related Phage Operator Sequences Validates Alignment Algorithm and Highlights the Importance of Overlooked Bonds. Genes. 2023; 14(12):2221. https://doi.org/10.3390/genes14122221
Chicago/Turabian StyleSedhom, Jacklin, and Lee A. Solomon. 2023. "Lambda CI Binding to Related Phage Operator Sequences Validates Alignment Algorithm and Highlights the Importance of Overlooked Bonds" Genes 14, no. 12: 2221. https://doi.org/10.3390/genes14122221
APA StyleSedhom, J., & Solomon, L. A. (2023). Lambda CI Binding to Related Phage Operator Sequences Validates Alignment Algorithm and Highlights the Importance of Overlooked Bonds. Genes, 14(12), 2221. https://doi.org/10.3390/genes14122221






