Selective Sweeps in Cattle Genomes in Response to the Influence of Urbanization and Environmental Contamination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Location, Sample Collection, and Analysis
2.2. Heavy Metal Analysis of Hair Samples
2.3. Genotyping and Quality Control
2.4. Selection Signature Analysis
3. Results
3.1. Selection Signatures According to SSI Grouping
3.2. Functional Analysis of Urbanization Effects
3.3. Selection Signatures According to Heavy Metal Contamination Grouping
4. Discussion
4.1. Selection Sweeps Due to Urbanization
4.2. Selection Sweeps Due to Heavy Metal Contamination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delgado, C.L. Rising Consumption of Meat and Milk in Developing Countries Has Created a New Food Revolution. J. Nutr. 2003, 133, 3907S–3910S. [Google Scholar] [CrossRef] [PubMed]
- Groot, M.J.; van’t Hooft, K.E. The Hidden Effects of Dairy Farming on Public and Environmental Health in the Netherlands, India, Ethiopia, and Uganda, Considering the Use of Antibiotics and Other Agro-Chemicals. Front. Public Health 2016, 4, 12. [Google Scholar] [CrossRef]
- Purfield, D.C.; McParland, S.; Wall, E.; Berry, D.P. The Distribution of Runs of Homozygosity and Selection Signatures in Six Commercial Meat Sheep Breeds. PLoS ONE 2017, 12, e0176780. [Google Scholar] [CrossRef] [PubMed]
- Eydivandi, S.; Roudbar, M.A.; Ardestani, S.S.; Momen, M.; Sahana, G. A Selection Signatures Study among Middle Eastern and European Sheep Breeds. J. Anim. Breed. Genet. 2021, 138, 574–588. [Google Scholar] [CrossRef]
- Fay, J.C.; Wu, C.-I. Hitchhiking Under Positive Darwinian Selection. Genetics 2000, 155, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.P.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting Recent Positive Selection in the Human Genome from Haplotype Structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A Map of Recent Positive Selection in the Human Genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Tang, K.; Thornton, K.R.; Stoneking, M. A New Approach for Using Genome Scans to Detect Recent Positive Selection in the Human Genome. PLoS Biol. 2007, 5, e171. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Schaffner, S.F.; Fry, B.; Lohmueller, J.; Varilly, P.; Shamovsky, O.; Palma, A.; Mikkelsen, T.S.; Altshuler, D.; Lander, E.S. Positive Natural Selection in the Human Lineage. Science 2006, 312, 1614–1620. [Google Scholar] [CrossRef]
- Reichenbach, M.; Pinto, A.; Malik, P.K.; Bhatta, R.; König, S.; Schlecht, E. Dairy Feed Efficiency and Urbanization–A System Approach in the Rural-Urban Interface of Bengaluru, India. Livest. Sci. 2021, 253, 104718. [Google Scholar] [CrossRef]
- Pinto, A.; Yin, T.; Reichenbach, M.; Bhatta, R.; Schlecht, E.; König, S. Phenotypic Dairy Cattle Trait Expressions in Dependency of Social-Ecological Characteristics along Rural–Urban Gradients. Sustainability 2020, 12, 9021. [Google Scholar] [CrossRef]
- Mullakkalparambil Velayudhan, S.; Brügemann, K.; Pinto, A.; Yin, T.; Reichenbach, M.; Sejian, V.; Bhatta, R.; Schlecht, E.; König, S. Effects of Heat Stress across the Rural-Urban Interface on Phenotypic Trait Expressions of Dairy Cattle in a Tropical Savanna Region. Sustainability 2022, 14, 4590. [Google Scholar] [CrossRef]
- Singh, M.; Ranvir, S.; Sharma, R.; Gandhi, K.; Mann, B. Assessment of Contamination of Milk and Milk Products with Heavy Metals. Indian J. Dairy Sci. 2020, 72, 608–615. [Google Scholar] [CrossRef]
- Yasotha, A.; Dabadé, D.S.; Singh, V.P.; Sivakumar, T. Risk Assessment of Heavy Metals in Milk from Cows Reared around Industrial Areas in India. Env. Geochem. Health 2021, 43, 1799–1815. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Silpa, M.V. Impacts of Heavy Metal Feed Contaminants in Cattle Farming. In Transfer of Mitigation Technologies for Heat Stress in Farm Animals; Seijian, V., Chauhan, S.S., Malik, P.K., Krishnan, G., Bagath, M., Kolte, A.P., Bhatta, R., Eds.; NIANP: Bengaluru, India, 2020; pp. 147–152. [Google Scholar]
- Alam, M.S.; Velayudhan, S.M.; Dey, D.K.; Adilieme, C.; Malik, P.K.; Bhatta, R.; König, S.; Schlecht, E. Urbanisation Threats to Dairy Cattle Health: Insights from Greater Bengaluru, India. Trop. Anim. Health Prod. 2023, 55, 350. [Google Scholar] [CrossRef] [PubMed]
- Varalakshmi, L.R.; Ganeshamurthy, A.N. Heavy Metal Contamination of Water Bodies, Soils and Vegetables in Peri Urban Areas of Bangalore City of India. In Proceedings of the 19th World Congress of Soil Science, Soil Solutions for a Changing World, Brisbane, Australia, 1–6 August 2010; pp. 1–6. [Google Scholar]
- Alam, M.S.; Schlecht, E.; Reichenbach, M. Impacts of COVID-19 on Small-Scale Dairy Enterprises in an Indian Megacity—Insights from Greater Bengaluru. Sustainability 2022, 14, 2057. [Google Scholar] [CrossRef]
- Reichenbach, M.; Pinto, A.; König, S.; Bhatta, R.; Schlecht, E. Dairy Production in an Urbanizing Environment—Typology and Linkages in the Megacity of Bengaluru, India. PLoS ONE 2021, 16, e0255791. [Google Scholar] [CrossRef]
- Velayudhan, S.M.; Brügemann, K.; Alam, S.; Yin, T.; Devaraj, C.; Sejian, V.; Schlecht, E.; König, S. Molecular, Physiological and Hematological Responses of Crossbred Dairy Cattle in a Tropical Savanna Climate. Biology 2022, 12, 26. [Google Scholar] [CrossRef]
- Hoffmann, E.; Jose, M.; Nölke, N.; Möckel, T. Construction and Use of a Simple Index of Urbanisation in the Rural–Urban Interface of Bangalore, India. Sustainability 2017, 9, 2146. [Google Scholar] [CrossRef]
- Kabata-Pendias, A.; Pendias, H. Trace Elements in Soils and Plants; CRC: Boca Raton, FL, USA, 1984. [Google Scholar]
- Pescod, M.B. Waste Water Treatment and Use in Agriculture; FAO: Rome, Italy, 1992. [Google Scholar]
- Awasthi, S.K. Prevention of Food Adulteration Act No 37 of 1954; Central and State Rules as Amended for 1999; Ashoka Law House: New Delhi, India, 2000.
- European Union. Commission Regulation (EC) No. 1881/2006 of 19 December Setting Maximum Levels for Certain Ontaminants in Foodstuffs. Off. J. Eur. Union 2006, 364, 5–24. [Google Scholar]
- BIS (Bureau of Indian Standards). Drinking Water Specification; BIS: New Delhi, India, 2012.
- IAEA International Atomic Energy Agency. Proceedings Series; Blix, H., Ed.; IAEA: Vienna, Austria, 1979; pp. 545–561. ISBN 92-0-010079-1. [Google Scholar]
- Paar, A. Application Collection. Document Number: E38IA046EN-A, Anton Paar GmbH, Germany; Document number: E38IA046EN-A; Anton Paar GmbH: Ostfildern, Germany, 2020. [Google Scholar]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M.; Vitalis, R. rehh: An R Package to Detect Footprints of Selection in Genome-Wide SNP Data from Haplotype Structure. Bioinformatics 2012, 28, 1176–1177. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M.; Klassmann, A.; Vitalis, R. rehh 2.0: A Reimplementation of the R Package rehh to Detect Positive Selection from Haplotype Structure. Mol. Ecol. Resour. 2017, 17, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Naderi, S.; Moradi, M.H.; Farhadian, M.; Yin, T.; Jaeger, M.; Scheper, C.; Korkuc, P.; Brockmann, G.A.; König, S.; May, K. Assessing selection signatures within and between selected lines of dual-purpose black and white and German Holstein cattle. Anim. Genet. 2020, 51, 391–408. [Google Scholar] [CrossRef]
- FAO. Mapping Supply and Demand for Animal-Source Foods to 2030; Robinson, T.P., Pozzi, F., Eds.; FAO: Rome, Italy, 2011. [Google Scholar]
- Marshall, K.; Gibson, J.P.; Mwai, O.; Mwacharo, J.M.; Haile, A.; Getachew, T.; Mrode, R.; Kemp, S.J. Livestock Genomics for Developing Countries–African Examples in Practice. Front. Genet. 2019, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kraft, P.; Nan, H.; Guo, Q.; Chen, C.; Qureshi, A.; Hankinson, S.E.; Hu, F.B.; Duffy, D.L.; Zhao, Z.Z.; et al. A Genome-Wide Association Study Identifies Novel Alleles Associated with Hair Color and Skin Pigmentation. PLoS Genet. 2008, 4, e1000074. [Google Scholar] [CrossRef]
- Ariyarathne, H.B.P.C.; Correa-Luna, M.; Blair, H.T.; Garrick, D.J.; Lopez-Villalobos, N. Identification of Genomic Regions Associated with Concentrations of Milk Fat, Protein, Urea and Efficiency of Crude Protein Utilization in Grazing Dairy Cows. Genes 2021, 12, 456. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, Z.; Zhang, Q.; Yue, S.; Yin, B.; Jiang, Y.; Shi, K. Identification of Whole-genome Significant Single Nucleotide Polymorphisms in Candidate Genes Associated with Body Conformation Traits in Chinese Holstein Cattle. Anim. Genet. 2020, 51, 141–146. [Google Scholar] [CrossRef]
- Nayeri, S.; Sargolzaei, M.; Abo-Ismail, M.K.; May, N.; Miller, S.P.; Schenkel, F.; Moore, S.S.; Stothard, P. Genome-Wide Association for Milk Production and Female Fertility Traits in Canadian Dairy Holstein Cattle. BMC Genet. 2016, 17, 75. [Google Scholar] [CrossRef]
- Maiorano, A.M.; Lourenco, D.L.; Tsuruta, S.; Ospina, A.M.T.; Stafuzza, N.B.; Masuda, Y.; Filho, A.E.V.; dos Cyrillo, J.N.S.G.; Curi, R.A.; de Silva, J.A.I.V. Assessing Genetic Architecture and Signatures of Selection of Dual Purpose Gir Cattle Populations Using Genomic Information. PLoS ONE 2018, 13, e0200694. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Raza, S.H.A.; Deng, J.; Ma, J.; Qu, X.; Yu, S.; Zhang, D.; Alshammari, A.M.; Almohaimeed, H.M.; et al. Identification of the Hub Genes Related to Adipose Tissue Metabolism of Bovine. Front. Vet. Sci. 2022, 9, 1014286. [Google Scholar] [CrossRef] [PubMed]
- Vallet, M.; Sophocleous, A.; Törnqvist, A.E.; Azfer, A.; Hof, R.V.T.; Albagha, O.M.; Ralston, S.H. Targeted Inactivation of Rin3 Increases Trabecular Bone Mass by Reducing Bone Resorption and Favouring Bone Formation. Calcif. Tissue Int. 2021, 109, 92–102. [Google Scholar] [CrossRef]
- Stronen, A.V.; Pertoldi, C.; Iacolina, L.; Kadarmideen, H.N.; Kristensen, T.N. Genomic Analyses Suggest Adaptive Differentiation of Northern European Native Cattle Breeds. Evol. Appl. 2019, 12, 1096–1113. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Castillero, M.; Then, C.; Altarriba, J.; Srihi, H.; López-Carbonell, D.; Díaz, C.; Martinez, P.; Hermida, M.; Varona, L. Detection of Genomic Regions with Pleiotropic Effects for Growth and Carcass Quality Traits in the Rubia Gallega Cattle Breed. Animals 2021, 11, 1682. [Google Scholar] [CrossRef]
- Bolormaa, S.; Hayes, B.J.; Savin, K.; Hawken, R.; Barendse, W.; Arthur, P.F.; Herd, R.M.; Goddard, M.E. Genome-Wide Association Studies for Feedlot and Growth Traits in Cattle1. J. Anim. Sci. 2011, 89, 1684–1697. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide Scan of Selection Signatures in Dehong Humped Cattle for Heat Tolerance and Disease Resistance. Anim. Genet. 2020, 51, 292–299. [Google Scholar] [CrossRef]
- Patel, P.; Raju, N.J.; Reddy, B.C.S.R.; Suresh, U.; Sankar, D.B.; Reddy, T.V.K. Heavy Metal Contamination in River Water and Sediments of the Swarnamukhi River Basin, India: Risk Assessment and Environmental Implications. Env. Geochem. Health 2018, 40, 609–623. [Google Scholar] [CrossRef]
- Hamsa, N.; Prakash, N.B. Heavy Metal Contamination in Soils and Crops Irrigated with Lakes of Bengaluru. Curr. Sci. 2020, 119, 1849. [Google Scholar] [CrossRef]
- Sundar, K.; Mukherjee, A.; Sadiq, M.; Chandrasekaran, N. Cr (III) Bioremoval Capacities of Indigenous and Adapted Bacterial Strains from Palar River Basin. J. Hazard. Mater. 2011, 187, 553–561. [Google Scholar] [CrossRef]
- Makridis, C.; Svarnas, C.; Rigas, N.; Gougoulias, N.; Roka, L.; Leontopoulos, S. Transfer of Heavy Metal Contaminants from Animal Feed to Animal Products. J. Agric. Sci. Technol. A 2012, 2, 149–154. [Google Scholar]
- Li, Y.; McCrory, D.F.; Powell, J.M.; Saam, H.; Jackson-Smith, D. A Survey of Selected Heavy Metal Concentrations in Wisconsin Dairy Feeds. J. Dairy Sci. 2005, 88, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Pilarczyk, R.; Wójcik, J.; Czerniak, P.; Sablik, P.; Pilarczyk, B.; Tomza-Marciniak, A. Concentrations of Toxic Heavy Metals and Trace Elements in Raw Milk of Simmental and Holstein-Friesian Cows from Organic Farm. Env. Monit. Assess. 2013, 185, 8383–8392. [Google Scholar] [CrossRef] [PubMed]
- Denholm, S.J.; Sneddon, A.A.; McNeilly, T.N.; Bashir, S.; Mitchell, M.C.; Wall, E. Phenotypic and Genetic Analysis of Milk and Serum Element Concentrations in Dairy Cows. J. Dairy Sci. 2019, 102, 11180–11192. [Google Scholar] [CrossRef] [PubMed]
- Flori, L.; Fritz, S.; Jaffrézic, F.; Boussaha, M.; Gut, I.; Heath, S.; Foulley, J.-L.; Gautier, M. The Genome Response to Artificial Selection: A Case Study in Dairy Cattle. PLoS ONE 2009, 4, e6595. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Dotsev, A.V.; Sermyagin, A.A.; Deniskova, T.E.; Abdelmanova, A.S.; Kharzinova, V.R.; Sölkner, J.; Reyer, H.; Wimmers, K.; Brem, G. Selection Signatures in Two Oldest Russian Native Cattle Breeds Revealed Using High-Density Single Nucleotide Polymorphism Analysis. PLoS ONE 2020, 15, e0242200. [Google Scholar] [CrossRef]
- Shi, D.; Dai, C.; Qin, J.; Gu, W. Negative Regulation of the P300-P53 Interplay by DDX24. Oncogene 2016, 35, 528–536. [Google Scholar] [CrossRef]
- DDX24 Gene-DEAD-Box Helicase 24. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DDX24&keywords=ddx24,bos,taurus (accessed on 16 September 2023).
- Millau, J.-F.; Bastien, N.; Drouin, R. P53 Transcriptional Activities: A General Overview and Some Thoughts. Mutat. Res. Rev. Mutat. Res. 2009, 681, 118–133. [Google Scholar] [CrossRef]
- Moore, S.G.; Pryce, J.E.; Hayes, B.J.; Chamberlain, A.J.; Kemper, K.E.; Berry, D.P.; McCabe, M.; Cormican, P.; Lonergan, P.; Fair, T.; et al. Differentially Expressed Genes in Endometrium and Corpus Luteum of Holstein Cows Selected for High and Low Fertility Are Enriched for Sequence Variants Associated with Fertility. Biol. Reprod. 2016, 94, 1–11. [Google Scholar] [CrossRef]
- Costa, A.; Lopez-Villalobos, N.; Visentin, G.; Marchi, M.D.; Cassandro, M.; Penasa, M. Heritability and Repeatability of Milk Lactose and Its Relationships with Traditional Milk Traits, Somatic Cell Score and Freezing Point in Holstein Cows. Animal 2019, 13, 909–916. [Google Scholar] [CrossRef]
- Doyle, J.L.; Berry, D.P.; Veerkamp, R.F.; Carthy, T.R.; Walsh, S.W.; Evans, R.D.; Purfield, D.C. Genomic Regions Associated With Skeletal Type Traits in Beef and Dairy Cattle Are Common to Regions Associated With Carcass Traits, Feed Intake and Calving Difficulty. Front. Genet. 2020, 11, 20. [Google Scholar] [CrossRef]
- Silva, R.P.D. Genomic Selection and Genome-Wide Association Study with Carcass Composition Indicator Traits in Nellore Cattle. Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brazil, 2021. [Google Scholar]
- Zhao, G.; Liu, Y.; Niu, Q.; Zheng, X.; Zhang, T.; Wang, Z.; Xu, L.; Zhu, B.; Gao, X.; Zhang, L.; et al. Runs of Homozygosity Analysis Reveals Consensus Homozygous Regions Affecting Production Traits in Chinese Simmental Beef Cattle. BMC Genom. 2021, 22, 678. [Google Scholar] [CrossRef] [PubMed]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-Wide Association Study on Milk Production and Somatic Cell Score for Thai Dairy Cattle Using Weighted Single-Step Approach with Random Regression Test-Day Model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Peters, S.O.; Akwanji, K.A.; Imumorin, I.G.; Zhao, X. High Density Genome Wide Genotyping-by-Sequencing and Association Identifies Common and Low Frequency SNPs, and Novel Candidate Genes Influencing Cow Milk Traits. Sci. Rep. 2016, 6, 31109. [Google Scholar] [CrossRef]
- Jiang, J.; Gao, Y.; Hou, Y.; Li, W.; Zhang, S.; Zhang, Q.; Sun, D. Whole-Genome Resequencing of Holstein Bulls for Indel Discovery and Identification of Genes Associated with Milk Composition Traits in Dairy Cattle. PLoS ONE 2016, 11, e0168946. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Wang, D.; Zhao, C.; Zhang, X.; Chen, Z.; Liu, J.; Sun, D.; Tang, H.; Wang, W.; Li, J.; et al. Longitudinal Genome-Wide Association Studies of Milk Production Traits in Holstein Cattle Using Whole-Genome Sequence Data Imputed from Medium-Density Chip Data. J. Dairy Sci. 2023, 106, 2535–2550. [Google Scholar] [CrossRef]
- Tahir, M.S.; Porto-Neto, L.R.; Gondro, C.; Shittu, O.B.; Wockner, K.; Tan, A.W.L.; Smith, H.R.; Gouveia, G.C.; Kour, J.; Fortes, M.R.S. Meta-Analysis of Heifer Traits Identified Reproductive Pathways in Bos Indicus Cattle. Genes 2021, 12, 768. [Google Scholar] [CrossRef]
- Livernois, A.M.; Mallard, B.A.; Cartwright, S.L.; Cánovas, A. Heat Stress and Immune Response Phenotype Affect DNA Methylation in Blood Mononuclear Cells from Holstein Dairy Cows. Sci. Rep. 2021, 11, 11371. [Google Scholar] [CrossRef]
- Mattmiller, S.A.; Corl, C.M.; Gandy, J.C.; Loor, J.J.; Sordillo, L.M. Glucose Transporter and Hypoxia-Associated Gene Expression in the Mammary Gland of Transition Dairy Cattle. J. Dairy Sci. 2011, 94, 2912–2922. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.; Lin, X.; Zhang, J.; Hou, G.; Zhang, L.; Liu, D.; Li, Y.; Li, J.; Xu, L. Genomic Inbreeding and Runs of Homozygosity Analysis of Indigenous Cattle Populations in Southern China. PLoS ONE 2022, 17, e0271718. [Google Scholar] [CrossRef]
- Seo, J.; Osorio, J.S.; Schmitt, E.; Corrêa, M.N.; Bertoni, G.; Trevisi, E.; Loor, J.J. Hepatic Purinergic Signaling Gene Network Expression and Its Relationship with Inflammation and Oxidative Stress Biomarkers in Blood from Peripartal Dairy Cattle. J. Dairy Sci. 2014, 97, 861–873. [Google Scholar] [CrossRef] [PubMed]
- May, K.; Weimann, C.; Scheper, C.; Strube, C.; König, S. Allele Substitution and Dominance Effects of CD166/ALCAM Gene Polymorphisms for Endoparasite Resistance and Test-Day Traits in a Small Cattle Population Using Logistic Regression Analyses. Mamm. Genome 2019, 30, 301–317. [Google Scholar] [CrossRef] [PubMed]
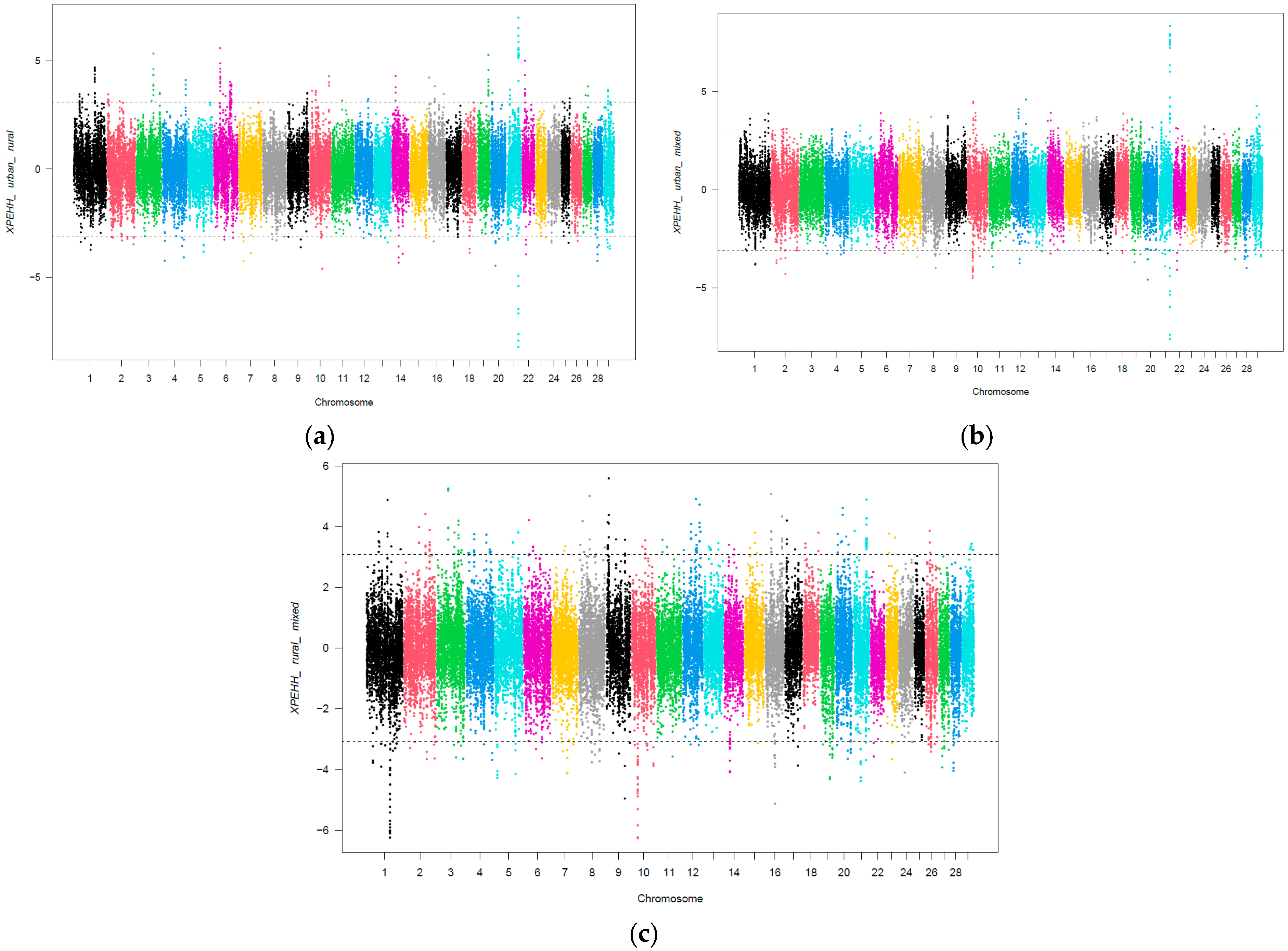
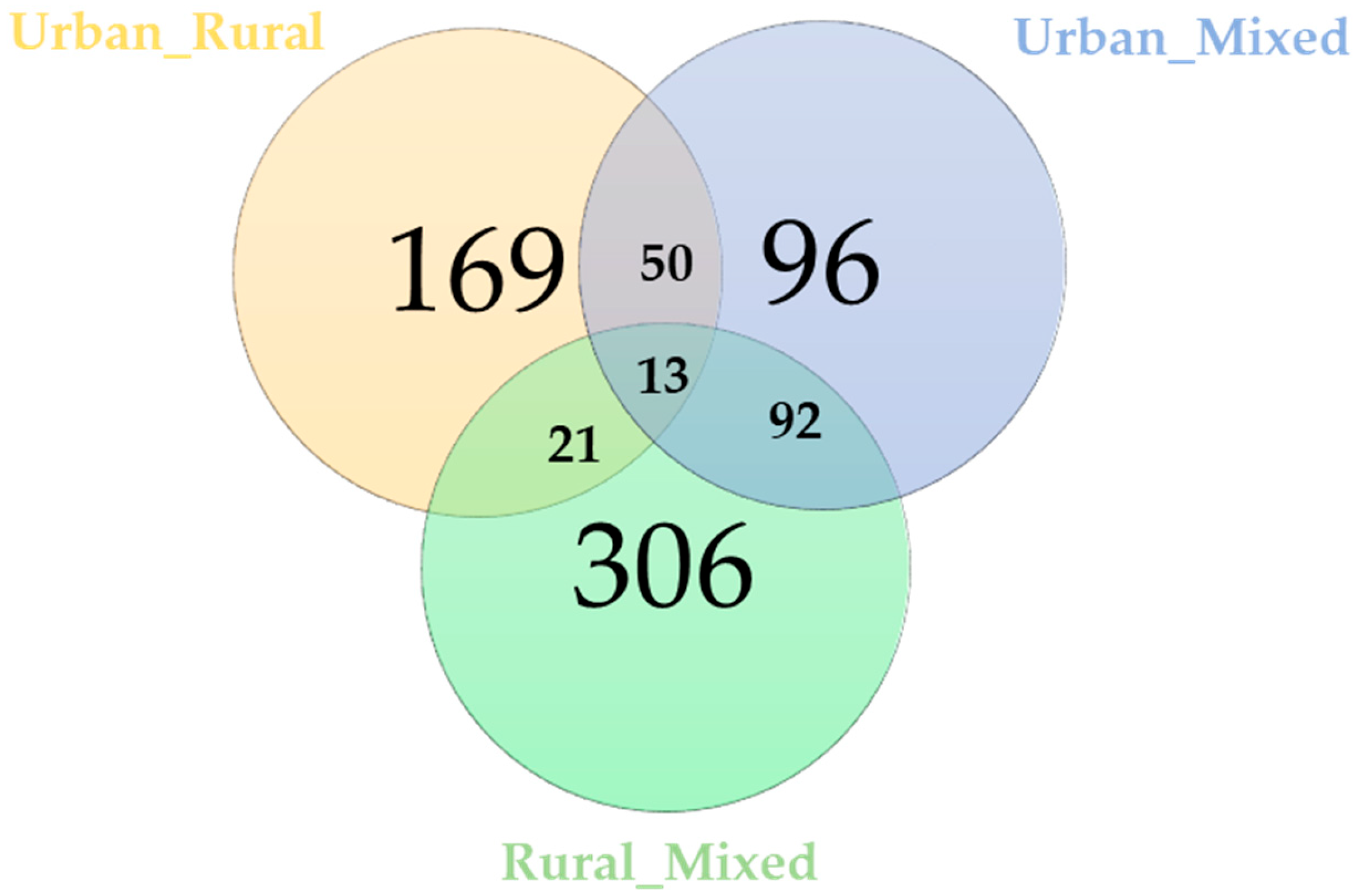
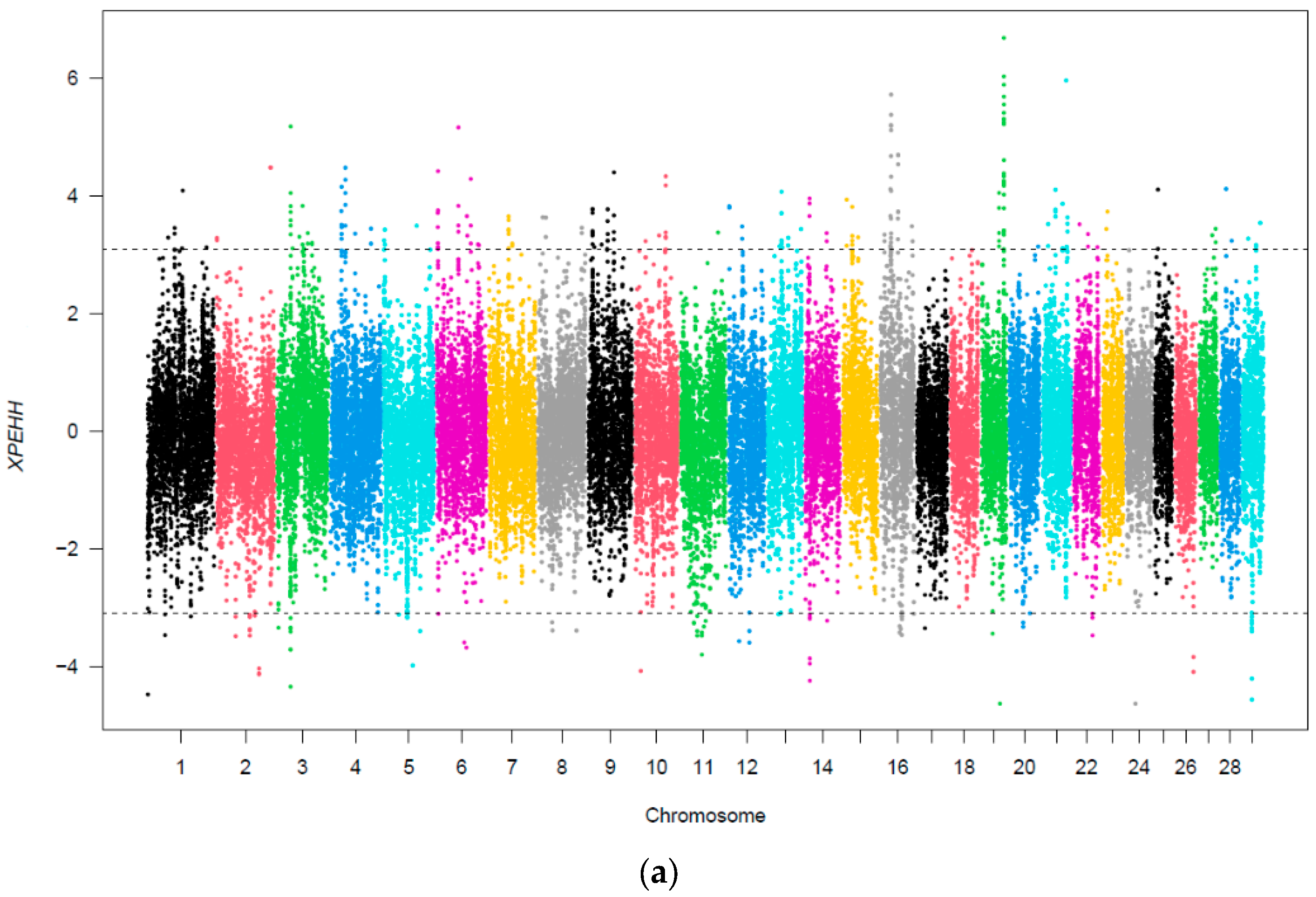

| Comparison | Gene Details |
|---|---|
| Urban_vs_Rural Positive (top 0.1 percentile) |
|
| Urban_vs_Rural Negative (bottom 0.1 percentile) |
|
| Urban_vs_Mixed Positive (top 0.1 percentile) |
|
| Urban_vs_Mixed Negative (bottom 0.1 percentile) |
|
| Rural_vs_Mixed Positive (top 0.1 percentile) |
|
| Rural_vs_Mixed Negative (bottom 0.1 percentile) |
|
| KEGG Term | Gene Count | Raw p-Value | Fold Enrichment |
|---|---|---|---|
| Urban_vs_Rural (Enrichment Score: 1.67) | |||
| bta00480: Glutathione metabolism | 4 | 0.01 | 10.92 |
| bta00982: Drug metabolism—cytochrome P450 | 4 | 0.01 | 10.92 |
| bta05204: Chemical carcinogenesis—DNA adducts | 4 | 0.01 | 10.41 |
| bta00980: Metabolism of xenobiotics by cytochrome P450 | 4 | 0.01 | 10.10 |
| bta01524: Platinum drug resistance | 4 | 0.01 | 8.68 |
| bta00983: Drug metabolism—other enzymes | 5 | 9.54 × 10−4 | 11.13 |
| bta05418: Fluid shear stress and atherosclerosis | 5 | 0.01 | 5.83 |
| bta05207: Chemical carcinogenesis—receptor activation | 5 | 0.04 | 3.92 |
| bta05208: Chemical carcinogenesis—reactive oxygen species | 5 | 0.05 | 3.53 |
| Urban_vs_Mixed (Enrichment Score: 0.95) | |||
| bta04913: Ovarian steroidogenesis | 3 | 0.04 | 8.69 |
| bta04927: Cortisol synthesis and secretion | 3 | 0.05 | 8.17 |
| Rural_vs_Mixed (Enrichment Score: 1.96) | |||
| bta04934: Cushing syndrome | 8 | 0.00 | 4.67 |
| bta04927: Cortisol synthesis and secretion | 5 | 0.01 | 6.88 |
| bta04917: Prolactin signaling pathway | 5 | 0.01 | 5.48 |
| bta04913: Ovarian steroidogenesis | 4 | 0.03 | 5.85 |
| GO Category | GO Term | Gene Count | Raw p-Value | Fold Enrichment |
|---|---|---|---|---|
| Cadmium (control_vs_treatment) | ||||
| Molecular Function | GO:0016887: ATPase activity | 7 | 0.03 | 3.02 |
| GO:0003777: Microtubule motor activity | 3 | 0.04 | 8.84 | |
| Cellular Component | GO:0005871: Kinesin complex | 3 | 0.04 | 9.14 |
| Lead (control_vs_treatment) | ||||
| Biological Process | GO:0007156: Homophilic cell adhesion via plasma membrane adhesion molecules | 11 | 3.27 × 10−8 | 11.68 |
| GO:0007155: Cell adhesion | 11 | 1.18 × 10−4 | 4.69 | |
| Molecular Function | GO:0005509: Calcium ion binding | 12 | 0.00 | 2.82 |
| Cellular Component | GO:0005887: Integral component of plasma membrane | 19 | 3.53 × 10−5 | 3.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velayudhan, S.M.; Alam, S.; Yin, T.; Brügemann, K.; Buerkert, A.; Sejian, V.; Bhatta, R.; Schlecht, E.; König, S. Selective Sweeps in Cattle Genomes in Response to the Influence of Urbanization and Environmental Contamination. Genes 2023, 14, 2083. https://doi.org/10.3390/genes14112083
Velayudhan SM, Alam S, Yin T, Brügemann K, Buerkert A, Sejian V, Bhatta R, Schlecht E, König S. Selective Sweeps in Cattle Genomes in Response to the Influence of Urbanization and Environmental Contamination. Genes. 2023; 14(11):2083. https://doi.org/10.3390/genes14112083
Chicago/Turabian StyleVelayudhan, Silpa Mullakkalparambil, Shahin Alam, Tong Yin, Kerstin Brügemann, Andreas Buerkert, Veerasamy Sejian, Raghavendra Bhatta, Eva Schlecht, and Sven König. 2023. "Selective Sweeps in Cattle Genomes in Response to the Influence of Urbanization and Environmental Contamination" Genes 14, no. 11: 2083. https://doi.org/10.3390/genes14112083
APA StyleVelayudhan, S. M., Alam, S., Yin, T., Brügemann, K., Buerkert, A., Sejian, V., Bhatta, R., Schlecht, E., & König, S. (2023). Selective Sweeps in Cattle Genomes in Response to the Influence of Urbanization and Environmental Contamination. Genes, 14(11), 2083. https://doi.org/10.3390/genes14112083








