A Genome-Wide Identification and Comparative Analysis of the Heavy-Metal-Associated Gene Family in Cucurbitaceae Species and Their Role in Cucurbita pepo under Arsenic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of HMA Genes in Cucurbits
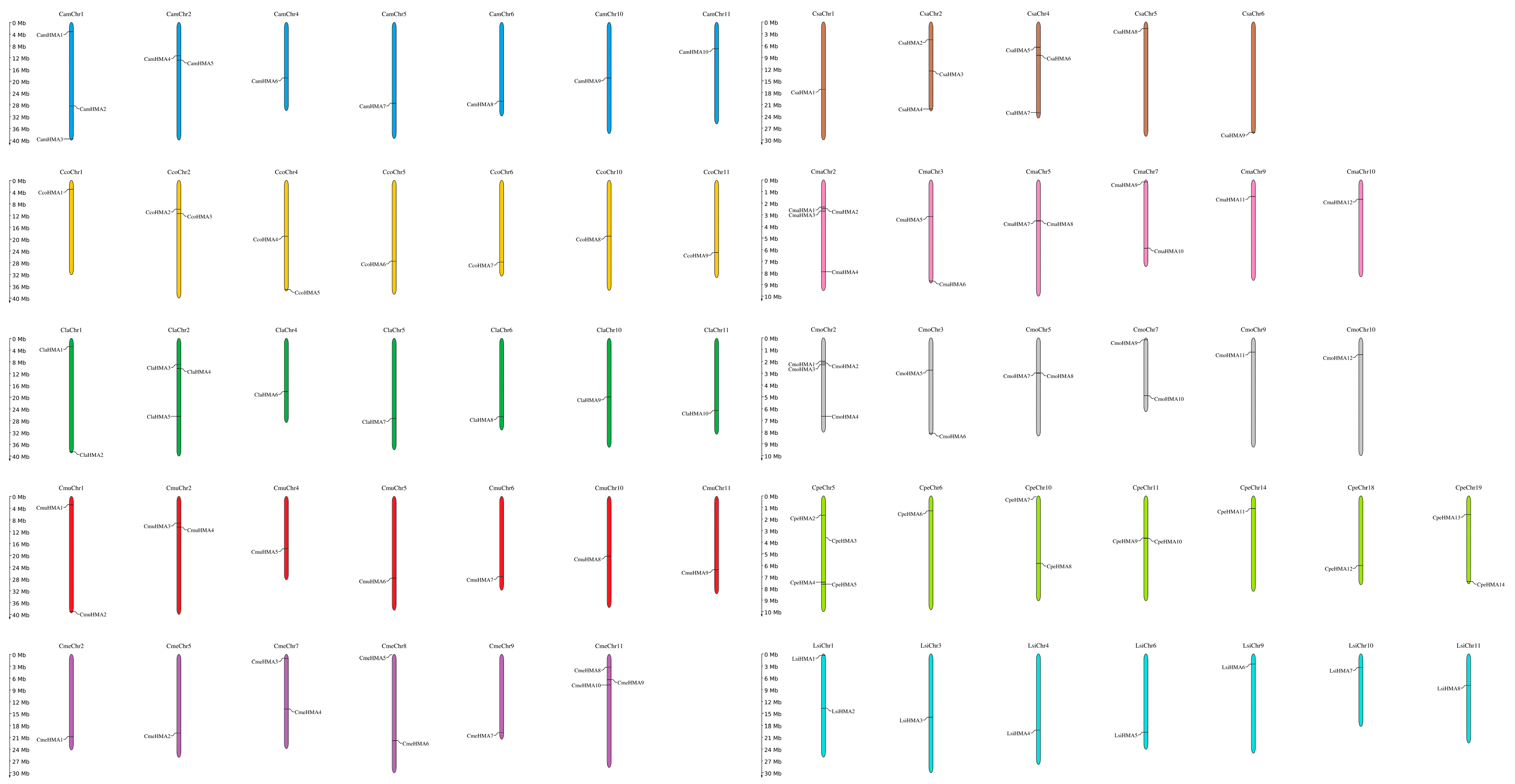
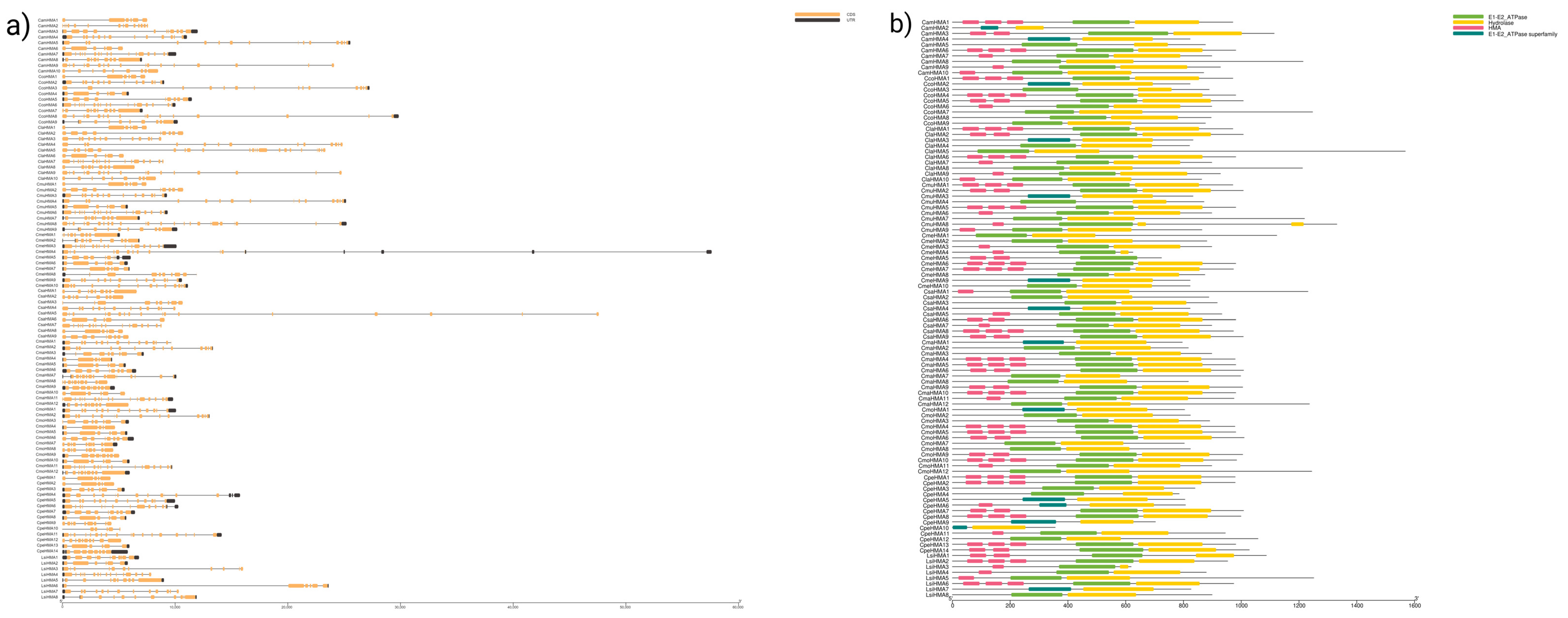
2.2. Chromosomal Location and Gene Structure of HMA Proteins
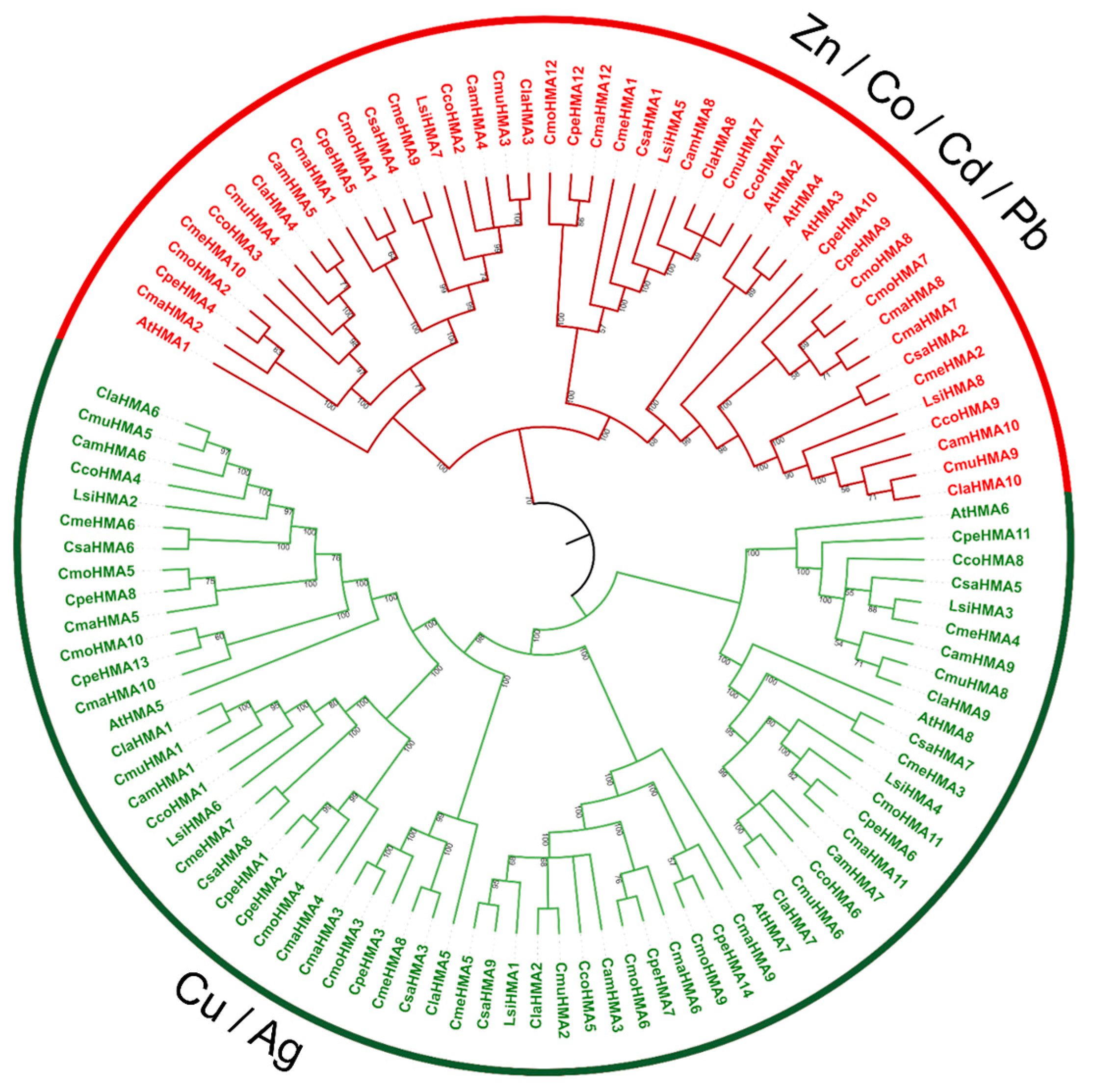
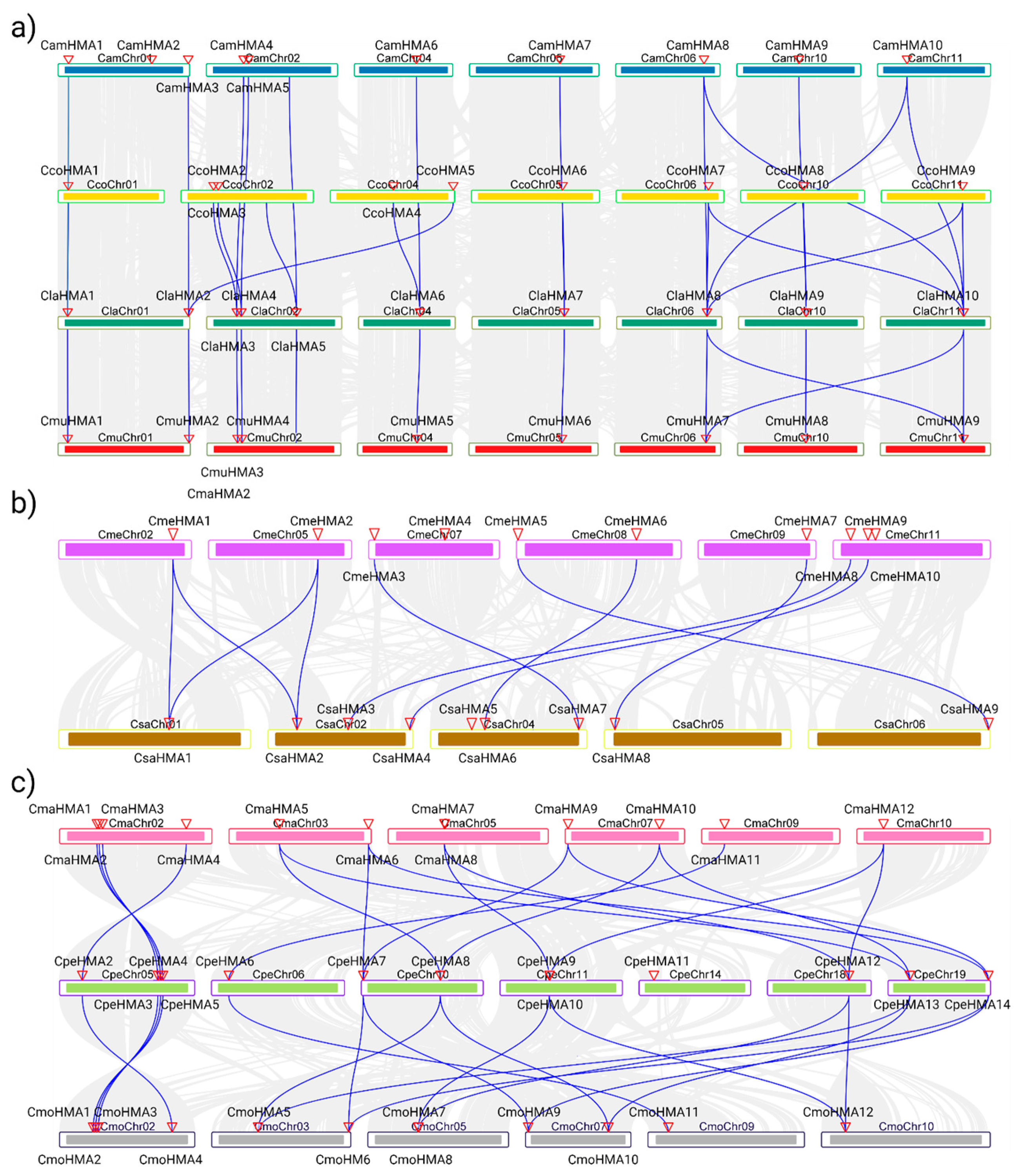
2.3. Phylogenetic Analysis, Synteny Analysis, and Gene Duplication Events of the Cucurbit HMA Family
2.4. Motif Analysis and Promoter Cis-Element Identification
2.5. Gene Ontology (GO) Annotation of HMA Proteins
2.6. Expression Pattern of the HMA Family in C. pepo under Different Cu Treatments
2.7. Expression Pattern with RT-qPCR of the HMA Family in Cucurbita pepo under As Treatment
3. Results
3.1. Identification of HMA Genes in Cucurbits
3.2. Chromosomal Location and Gene Structure of HMA Genes
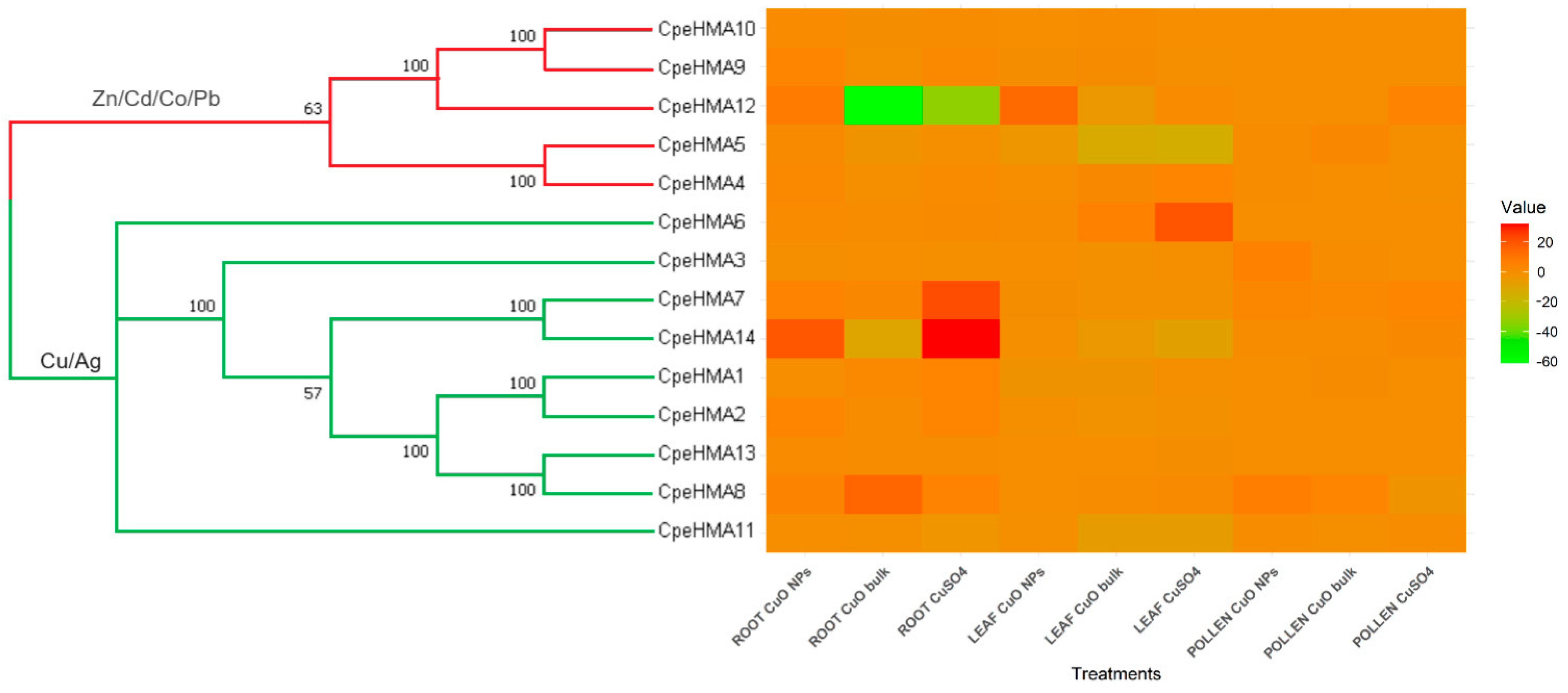
3.3. Phylogenetic Analysis, Synteny Analysis, and Gene Duplication Events of the Cucurbit HMA Gene Family
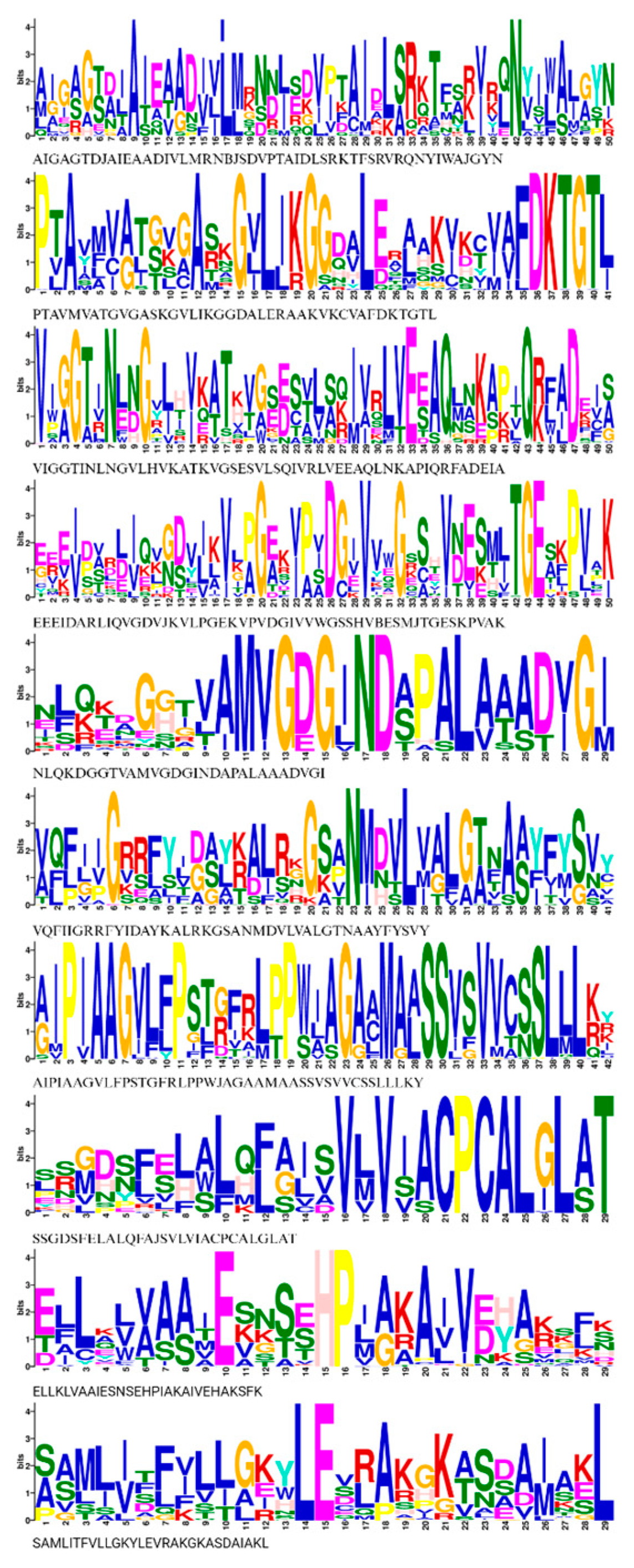
3.4. Conserved Motif Analysis and Cis-Elements of HMA Proteins
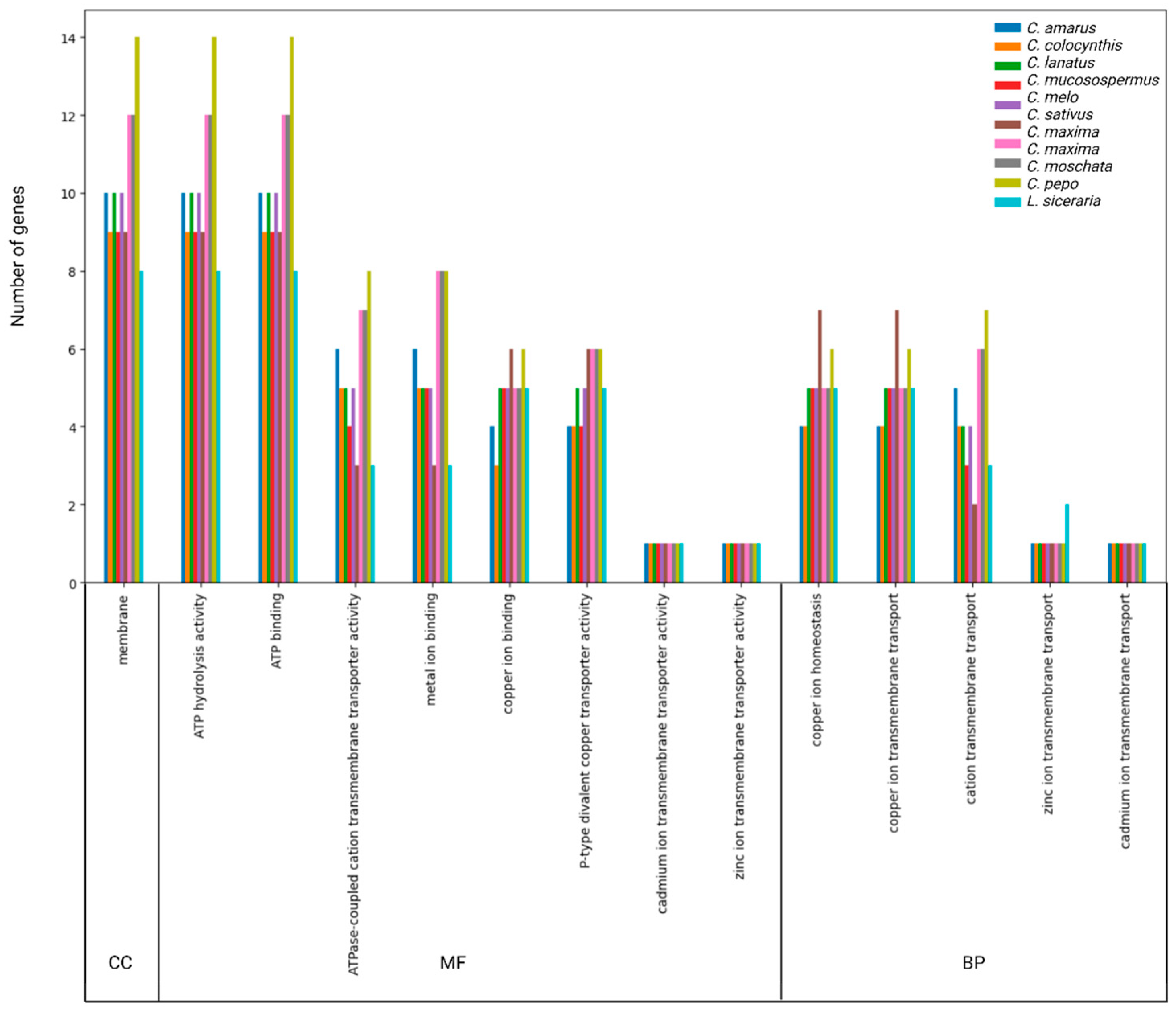
3.5. GO Annotation of HMA Proteins
3.6. Gene Expression Pattern of HMA Genes in Tissues of Cucurbita Pepo under Cu and as Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin and domestication of Cucurbitaceae crops: Insights from phylogenies, genomics and archaeology. New Phytol. 2020, 226, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; He, Z.; Chen, N.; Tang, Z.; Wang, Q.; Cai, Y. The roles of environmental factors in regulation of oxidative stress in plant. BioMed Res. Int. 2019, 2019, 9732325. [Google Scholar] [CrossRef] [PubMed]
- ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Han, Y.-H.; Cao, Y.; Zhu, Y.-G.; Rathinasabapathi, B.; Ma, L.Q. Arsenic transport in rice and biological solutions to reduce arsenic risk from rice. Front. Plant Sci. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Podgorski, J.; Berg, M. Global threat of arsenic in groundwater. Science 2020, 368, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Singla, P. Arsenic toxicity in crop plants: Physiological effects and tolerance mechanisms. Environ. Chem. Lett. 2011, 9, 303–321. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Smith, A.T.; Smith, K.P.; Rosenzweig, A.C. Diversity of the metal-transporting P 1B-type ATPases. JBIC J. Biol. Inorg. Chem. 2014, 19, 947–960. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Nissen, P. P-type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef]
- Mills, R.F.; Krijger, G.C.; Baccarini, P.J.; Hall, J.; Williams, L.E. Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J. 2003, 35, 164–176. [Google Scholar] [CrossRef]
- Gravot, A.; Lieutaud, A.; Verret, F.; Auroy, P.; Vavasseur, A.; Richaud, P. AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Liu, H.; Zheng, H.; Li, X.; Zhao, H.; Zou, D. Heavy metal stress-associated proteins in rice and Arabidopsis: Genome-wide identification, phylogenetics, duplication, and expression profiles analysis. Front. Genet. 2020, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Zorrig, W.; Abdelly, C.; Berthomieu, P. The phylogenetic tree gathering the plant Zn/Cd/Pb/Co P1B-ATPases appears to be structured according to the botanical families. Comptes Rendus Biol. 2011, 334, 863–871. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P1B-ATPases–an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Argüello, J.M.; Eren, E.; González-Guerrero, M. The structure and function of heavy metal transport P 1B-ATPases. Biometals 2007, 20, 233–248. [Google Scholar] [CrossRef]
- Axelsen, K.B.; Palmgren, M.G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998, 46, 84–101. [Google Scholar] [CrossRef]
- Takahashi, R.; Bashir, K.; Ishimaru, Y.; Nishizawa, N.K.; Nakanishi, H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 2012, 7, 1605–1607. [Google Scholar] [CrossRef]
- Li, D.; Xu, X.; Hu, X.; Liu, Q.; Wang, Z.; Zhang, H.; Wang, H.; Wei, M.; Wang, H.; Liu, H. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa. Front. Plant Sci. 2015, 6, 1149. [Google Scholar] [CrossRef]
- Fang, X.; Wang, L.; Deng, X.; Wang, P.; Ma, Q.; Nian, H.; Wang, Y.; Yang, C. Genome-wide characterization of soybean P 1B-ATPases gene family provides functional implications in cadmium responses. BMC Genom. 2016, 17, 1–15. [Google Scholar] [CrossRef]
- Zhiguo, E.; Tingting, L.; Chen, C.; Lei, W. Genome-wide survey and expression analysis of P1B-ATPases in rice, maize and sorghum. Rice Sci. 2018, 25, 208–217. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Q.; Zhang, X.; Zhang, X.; Yu, T.; Wu, Y.; Fang, Y.; Xue, D. Genome-wide identification of the HMA gene family and expression analysis under Cd stress in barley. Plants 2021, 10, 1849. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.F.; Peaston, K.A.; Runions, J.; Williams, L.E. HvHMA2, a P1B-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS ONE 2012, 7, e42640. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xiao, H.; Sun, J.; Wang, S.; Wang, J.; Chang, P.; Zhou, X.; Lei, B.; Lu, K.; Luo, F. Genome-wide analysis and expression profiling of the HMA gene family in Brassica napus under cd stress. Plant Soil 2018, 426, 365–381. [Google Scholar] [CrossRef]
- Manzoor, M.A.; Cheng, X.; Li, G.; Su, X.; Abdullah, M.; Cai, Y. Gene structure, evolution and expression analysis of the P-ATPase gene family in Chinese pear (Pyrus bretschneideri). Comput. Biol. Chem. 2020, 88, 107346. [Google Scholar] [CrossRef]
- Fan, W.; Liu, C.; Cao, B.; Qin, M.; Long, D.; Xiang, Z.; Zhao, A. Genome-wide identification and characterization of four gene families putatively involved in cadmium uptake, translocation and sequestration in mulberry. Front. Plant Sci. 2018, 9, 879. [Google Scholar] [CrossRef]
- Ma, Y.; Wei, N.; Wang, Q.; Liu, Z.; Liu, W. Genome-wide identification and characterization of the heavy metal ATPase (HMA) gene family in Medicago truncatula under copper stress. Int. J. Biol. Macromol. 2021, 193, 893–902. [Google Scholar] [CrossRef]
- Ye, X.; Liu, C.; Yan, H.; Wan, Y.; Wu, Q.; Wu, X.; Zhao, G.; Zou, L.; Xiang, D. Genome-wide identification and transcriptome analysis of the heavy metal-associated (HMA) gene family in Tartary buckwheat and their regulatory roles under cadmium stress. Gene 2022, 847, 146884. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Choi, H.; Segami, S.; Cho, H.T.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 contributes to the detoxification of excess Zn (II) in Arabidopsis. Plant J. 2009, 58, 737–753. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.J.; Xu, W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar] [CrossRef]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef]
- Feng, S.; Shen, Y.; Xu, H.; Dong, J.; Chen, K.; Xiang, Y.; Jiang, X.; Yao, C.; Lu, T.; Huan, W. RNA-Seq Identification of Cd Responsive Transporters Provides Insights into the Association of Oxidation Resistance and Cd Accumulation in Cucumis sativus L. Antioxidants 2021, 10, 1973. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Gao, Y.; Wu, L.; Wang, L.; Zhang, T.; Dai, C.; Xu, W.; Feng, L.; Ma, M.; Zhu, Y.-G. Potential use of the Pteris vittata arsenic hyperaccumulation-regulation network for phytoremediation. J. Hazard. Mater. 2019, 368, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Axelsen, K.B.; Palmgren, M.G. Inventory of the Superfamily of P-Type Ion Pumps in Arabidopsis. Plant Physiol. 2001, 126, 696–706. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Moniz de Sá, M.; Drouin, G. Phylogeny and substitution rates of angiosperm actin genes. Mol. Biol. Evol. 1996, 13, 1198–1212. [Google Scholar] [CrossRef] [PubMed]
- Marmiroli, M.; Pagano, L.; Rossi, R.; De La Torre-Roche, R.; Lepore, G.O.; Ruotolo, R.; Gariani, G.; Bonanni, V.; Pollastri, S.; Puri, A. Copper oxide nanomaterial fate in plant tissue: Nanoscale impacts on reproductive tissues. Environ. Sci. Technol. 2021, 55, 10769–10783. [Google Scholar] [CrossRef] [PubMed]
- Obrero, Á.; Die, J.V.; Román, B.; Gómez, P.; Nadal, S.; González-Verdejo, C.I. Selection of Reference Genes for Gene Expression Studies in Zucchini (Cucurbita pepo) Using qPCR. J. Agric. Food Chem. 2011, 59, 5402–5411. [Google Scholar] [CrossRef] [PubMed]
- Obrero, Á.; González-Verdejo, C.I.; Die, J.V.; Gómez, P.; Del Río-Celestino, M.; Román, B. Carotenogenic Gene Expression and Carotenoid Accumulation in Three Varieties of Cucurbita pepo during Fruit Development. J. Agric. Food Chem. 2013, 61, 6393–6403. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- David, R.; Byrt, C.S.; Tyerman, S.D.; Gilliham, M.; Wege, S. Roles of membrane transporters: Connecting the dots from sequence to phenotype. Ann. Bot. 2019, 124, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.E.; Jarvis, R.S.; Sherson, S.M.; Cobbett, C.S. Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 2009, 181, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, N.; Cun, P.; Richaud, P.; Vavasseur, A. Zn/Cd/Co/Pb P1B-ATPases in plants, physiological roles and biological interest. In Metal Toxicity in Plants: Perception, Signaling and Remediation; Springer: Berlin/Heidelberg, Germany, 2011; pp. 227–248. [Google Scholar]
- Wu, D.; Saleem, M.; He, T.; He, G. The mechanism of metal homeostasis in plants: A new view on the synergistic regulation pathway of membrane proteins, lipids and metal ions. Membranes 2021, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.T.; Barupala, D.; Stemmler, T.L.; Rosenzweig, A.C. A new metal binding domain involved in cadmium, cobalt and zinc transport. Nat. Chem. Biol. 2015, 11, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Molteni, E.; Huffman, D.L.; O’Halloran, T.V. Metallochaperones and Metal-Transporting ATPases: A Comparative Analysis of Sequences and Structures. Genome Res. 2002, 12, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, Y.; Lim, C.; Tosha, T.; Yoshida, K.; Hagai, T.; Akiyama, S.; Watanabe, S.; Nakagome, K.; Shiro, Y. Identification of a novel zinc-binding protein, C1orf123, as an interactor with a heavy metal-associated domain. PLoS ONE 2018, 13, e0204355. [Google Scholar] [CrossRef]
- Huang, Q.; Qiu, W.; Yu, M.; Li, S.; Lu, Z.; Zhu, Y.; Kan, X.; Zhuo, R. Genome-Wide Characterization of Sedum plumbizincicola HMA Gene Family Provides Functional Implications in Cadmium Response. Plants 2022, 11, 215. [Google Scholar]
- Abbas, G.; Murtaza, B.; Bibi, I.; Shahid, M.; Niazi, N.K.; Khan, M.I.; Amjad, M.; Hussain, M.; Natasha. Arsenic Uptake, Toxicity, Detoxification, and Speciation in Plants: Physiological, Biochemical, and Molecular Aspects. Int. J. Environ. Res. Public Health 2018, 15, 59. [Google Scholar] [CrossRef]
- Garbinski, L.D.; Rosen, B.P.; Chen, J. Pathways of arsenic uptake and efflux. Environ. Int. 2019, 126, 585–597. [Google Scholar] [CrossRef]
- Li, N.; Wang, J.; Song, W.-Y. Arsenic uptake and translocation in plants. Plant Cell Physiol. 2016, 57, 4–13. [Google Scholar] [CrossRef]
- Bali, A.S.; Sidhu, G.P.S. Arsenic acquisition, toxicity and tolerance in plants-From physiology to remediation: A review. Chemosphere 2021, 283, 131050. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Park, J.; Mendoza-Cózatl, D.G.; Suter-Grotemeyer, M.; Shim, D.; Hörtensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Yamaki, T.; Yamaji, N.; Ko, D.; Jung, K.-H.; Fujii-Kashino, M.; An, G.; Martinoia, E.; Lee, Y.; Ma, J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 2014, 111, 15699–15704. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Sakurai, G.; Mitani-Ueno, N.; Ma, J.F. Orchestration of three transporters and distinct vascular structures in node for intervascular transfer of silicon in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11401–11406. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, Y.; Chen, F.; Ji, Y.; Zhao, F.-J. OsPTR7 (OsNPF8. 1), a putative peptide transporter in rice, is involved in dimethylarsenate accumulation in rice grain. Plant Cell Physiol. 2017, 58, 904–913. [Google Scholar] [CrossRef]
- Mathews, S.; Rathinasabapathi, B.; Ma, L.Q. Uptake and translocation of arsenite by Pteris vittata L.: Effects of glycerol, antimonite and silver. Environ. Pollut. 2011, 159, 3490–3495. [Google Scholar] [CrossRef]
- Nagarajan, V.; Ebbs, S. Transport of arsenite by the arsenic hyperaccumulating brake fern Pteris vittata is inhibited by monovalent silver. Indian J. Plant Physiol. 2007, 12, 312–316. [Google Scholar]
- Mayerhofer, H.; Sautron, E.; Rolland, N.; Catty, P.; Seigneurin-Berny, D.; Pebay-Peyroula, E.; Ravaud, S. Structural Insights into the Nucleotide-Binding Domains of the P1B-type ATPases HMA6 and HMA8 from Arabidopsis thaliana. PLoS ONE 2016, 11, e0165666. [Google Scholar] [CrossRef]
- Sautron, E.; Giustini, C.; Dang, T.; Moyet, L.; Salvi, D.; Crouzy, S.; Rolland, N.; Catty, P.; Seigneurin-Berny, D. Identification of Two Conserved Residues Involved in Copper Release from Chloroplast PIB-1-ATPases *. J. Biol. Chem. 2016, 291, 20136–20148. [Google Scholar] [CrossRef]
- Sautron, E.; Mayerhofer, H.; Giustini, C.; Pro, D.; Crouzy, S.; Ravaud, S.; Pebay-Peyroula, E.; Rolland, N.; Catty, P.; Seigneurin-Berny, D. HMA6 and HMA8 are two chloroplast Cu+-ATPases with different enzymatic properties. Biosci. Rep. 2015, 35, e00201. [Google Scholar] [CrossRef]
- Shingles, R.; Wimmers, L.E.; McCarty, R.E. Copper transport across pea thylakoid membranes. Plant Physiol. 2004, 135, 145–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gresser, M.J. ADP-arsenate. Formation by submitochondrial particles under phosphorylating conditions. J. Biol. Chem. 1981, 256, 5981–5983. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kuroda, K.; Kimura, K.; Southron-Francis, J.L.; Furuzawa, A.; Kimura, K.; Iuchi, S.; Kobayashi, M.; Taylor, G.J.; Koyama, H. Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 2008, 148, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hao, C.; Du, J.; Xu, L.; Guo, Z.; Li, D.; Cai, H.; Guo, H.; Li, L. The carboxy terminal transmembrane domain of SPL7 mediates interaction with RAN1 at the endoplasmic reticulum to regulate ethylene signalling in Arabidopsis. New Phytol. 2022, 236, 878–892. [Google Scholar] [CrossRef]
- Freitas, E.O.; Melo, B.P.; Lourenço-Tessutti, I.T.; Arraes, F.B.M.; Amorim, R.M.; Lisei-de-Sá, M.E.; Costa, J.A.; Leite, A.G.B.; Faheem, M.; Ferreira, M.A.; et al. Identification and characterization of the GmRD26 soybean promoter in response to abiotic stresses: Potential tool for biotechnological application. BMC Biotechnol. 2019, 19, 79. [Google Scholar] [CrossRef]

| Species | HMA Clade | Total | |
|---|---|---|---|
| Zn/Cd/Co/Pb | Cu/Ag | ||
| Arabidopsis thaliana | 4 | 4 | 8 |
| Brassica napus | 17 | 14 | 31 |
| Citrullus amarus | 4 | 6 | 10 |
| Citrullus colocynthis | 4 | 5 | 9 |
| Citrullus lanatus | 4 | 6 | 10 |
| Citrullus mucusospermus | 4 | 5 | 9 |
| Cucumis melo | 4 | 6 | 10 |
| Cucumis sativus | 3 | 6 | 9 |
| Cucurbita maxima | 5 | 7 | 12 |
| Cucurbita moschata | 5 | 7 | 12 |
| Cucurbita pepo | 5 | 9 | 14 |
| Glycine max | 6 | 14 | 20 |
| Hordeum vulgare | 3 | 6 | 9 |
| Legenaria siceraria | 3 | 5 | 8 |
| Linum usitatissimum | 4 | 8 | 12 |
| Medicago truncatula | 2 | 7 | 9 |
| Morus alba | 2 | 6 | 8 |
| Oryza sativa | 3 | 6 | 9 |
| Populus trichocarpa | 2 | 10 | 12 |
| Pyrus bretschneideri | 1 | 7 | 8 |
| Sorghum bicolor | 4 | 7 | 11 |
| Fagopyrum tataricum | 2 | 5 | 7 |
| Zea mays | 5 | 6 | 11 |
| Species | Pair | Gene Names | Ka | Ks | Ka/Ks Ratio | Duplication Type | MYA 1 |
|---|---|---|---|---|---|---|---|
| C. amarus | 1 | CamHMA2-CamHMA3 | 0.5081 | 0.4684 | 1.0846 | Tandem | 35.70 |
| 2 | CamHMA1-CamHMA6 | 0.1036 | 0.4185 | 0.2474 | Segmental | 31.90 | |
| 3 | CamHMA8-CamHMA10 | 0.1208 | 0.3328 | 0.3629 | Segmental | 25.36 | |
| 4 | CamHMA7-CamHMA9 | 0.2087 | 0.4635 | 0.4503 | Segmental | 35.33 | |
| 5 | CamHMA4-CamHMA5 | 0.0796 | 0.2789 | 0.2855 | Tandem | 21.25 | |
| C. colocynthis | 1 | CcoHMA1-CcoHMA4 | 0.1056 | 0.4402 | 0.2400 | Segmental | 33.55 |
| 2 | CcoHMA7-CcoHMA9 | 0.1192 | 0.3347 | 0.3562 | Segmental | 25.51 | |
| 3 | CcoHMA6-CcoHMA8 | 0.2019 | 0.4616 | 0.4374 | Segmental | 35.18 | |
| 4 | CcoHMA2-CcoHMA3 | 0.0823 | 0.2832 | 0.2907 | Tandem | 21.59 | |
| C. lanatus | 1 | ClaHMA2-ClaHMA8 | 0.3042 | 0.4742 | 0.6415 | Segmental | 36.14 |
| 2 | ClaHMA1-ClaHMA6 | 0.1059 | 0.4301 | 0.2463 | Segmental | 32.78 | |
| 3 | ClaHMA5-ClaHMA10 | 0.1174 | 0.3248 | 0.3615 | Segmental | 24.75 | |
| 4 | ClaHMA7-ClaHMA9 | 0.2128 | 0.4653 | 0.4573 | Segmental | 35.46 | |
| 5 | ClaHMA3-ClaHMA4 | 0.0832 | 0.2848 | 0.2922 | Tandem | 21.70 | |
| C. mucusospermus | 1 | CmuHMA1-CmuHMA5 | 0.1075 | 0.4420 | 0.2431 | Segmental | 33.69 |
| 2 | CmuHMA7-CmuHMA9 | 0.1212 | 0.3426 | 0.3538 | Segmental | 26.11 | |
| 3 | CmuHMA6-CmuHMA8 | 0.2076 | 0.4521 | 0.4591 | Segmental | 34.46 | |
| 4 | CmuHMA3-CmuHMA4 | 0.0847 | 0.2817 | 0.3009 | Tandem | 21.47 | |
| C. melo | 1 | CmeHMA6-CmeHMA7 | 0.1115 | 0.4288 | 0.2599 | Segmental | 32.68 |
| 2 | CmeHMA2-CmeHMA4 | 0.2848 | 0.3743 | 0.7608 | Segmental | 28.52 | |
| 3 | CmeHMA9-CmeHMA10 | 0.0771 | 0.2719 | 0.2835 | Tandem | 20.72 | |
| 4 | CmeHMA3-CmeHMA5 | 0.2542 | 0.3687 | 0.6894 | Segmental | 28.10 | |
| C. sativus | 1 | CsaHMA6-CsaHMA8 | 0.1077 | 0.4426 | 0.2434 | Segmental | 33.73 |
| 2 | CsaHMA1-CsaHMA2 | 0.1225 | 0.3678 | 0.3330 | Segmental | 28.03 | |
| 3 | CsaHMA5-CsaHMA7 | 0.2109 | 0.4253 | 0.4958 | Segmental | 32.41 | |
| 4 | CsaHMA4-CsaHMA9 | 0.3201 | 0.4293 | 0.7455 | Segmental | 32.72 | |
| C. maxima | 1 | CmaHMA7-CmaHMA8 | 0.0256 | 0.0308 | 0.8303 | Tandem | 2.35 |
| 2 | CmaHMA6-CmaHMA9 | 0.0272 | 0.1394 | 0.1951 | Segmental | 10.62 | |
| 3 | CmaHMA1-CmaHMA2 | 0.0703 | 0.2765 | 0.2543 | Tandem | 21.07 | |
| 4 | CmaHMA3-CmaHMA11 | 0.2830 | 0.4255 | 0.6651 | Segmental | 32.43 | |
| 5 | CmaHMA5-CmaHMA10 | 0.0186 | 0.1056 | 0.1762 | Segmental | 8.05 | |
| C. moschata | 1 | CmoHMA3-CmoHMA11 | 0.3381 | 0.5363 | 0.6304 | Segmental | 40.88 |
| 2 | CmoHMA1-CmoHMA2 | 0.0874 | 0.3327 | 0.2628 | Tandem | 25.36 | |
| 3 | CmoHMA5-CmoHMA10 | 0.0190 | 0.1179 | 0.1612 | Segmental | 8.99 | |
| 4 | CmoHMA6-CmoHMA9 | 0.0241 | 0.1136 | 0.2125 | Segmental | 8.66 | |
| 5 | CmoHMA7-CmoHMA8 | 0.0130 | 0.0167 | 0.7802 | Tandem | 1.27 | |
| C. pepo | 1 | CpeHMA4-CpeHMA5 | 0.0981 | 0.3070 | 0.3196 | Tandem | 23.39 |
| 2 | CpeHMA7-CpeHMA14 | 0.0294 | 0.1337 | 0.2198 | Segmental | 10.19 | |
| 3 | CpeHMA6-CpeHMA11 | 0.2511 | 0.4202 | 0.5975 | Segmental | 32.03 | |
| 4 | CpeHMA9-CpeHMA10 | 0.0170 | 0.0301 | 0.5649 | Tandem | 2.29 | |
| 5 | CpeHMA8-CpeHMA13 | 0.0183 | 0.1176 | 0.1562 | Segmental | 8.96 | |
| L. siceraria | 1 | LsiHMA5-LsiHMA8 | 0.1290 | 0.3793 | 0.3400 | Segmental | 28.91 |
| 2 | LsiHMA3-LsiHMA4 | 0.2196 | 0.4498 | 0.4881 | Segmental | 34.28 | |
| 3 | LsiHMA1-LsiHMA7 | 0.2933 | 0.3786 | 0.7748 | Segmental | 28.85 | |
| 4 | LsiHMA2-LsiHMA6 | 0.0856 | 0.3274 | 0.2615 | Segmental | 24.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Iga, G.; Lopez-Ortiz, C.; Gracia-Rodriguez, C.; Almeida, A.; Nimmakayala, P.; Reddy, U.K.; Balagurusamy, N. A Genome-Wide Identification and Comparative Analysis of the Heavy-Metal-Associated Gene Family in Cucurbitaceae Species and Their Role in Cucurbita pepo under Arsenic Stress. Genes 2023, 14, 1877. https://doi.org/10.3390/genes14101877
Flores-Iga G, Lopez-Ortiz C, Gracia-Rodriguez C, Almeida A, Nimmakayala P, Reddy UK, Balagurusamy N. A Genome-Wide Identification and Comparative Analysis of the Heavy-Metal-Associated Gene Family in Cucurbitaceae Species and Their Role in Cucurbita pepo under Arsenic Stress. Genes. 2023; 14(10):1877. https://doi.org/10.3390/genes14101877
Chicago/Turabian StyleFlores-Iga, Gerardo, Carlos Lopez-Ortiz, Celeste Gracia-Rodriguez, Aldo Almeida, Padma Nimmakayala, Umesh K. Reddy, and Nagamani Balagurusamy. 2023. "A Genome-Wide Identification and Comparative Analysis of the Heavy-Metal-Associated Gene Family in Cucurbitaceae Species and Their Role in Cucurbita pepo under Arsenic Stress" Genes 14, no. 10: 1877. https://doi.org/10.3390/genes14101877
APA StyleFlores-Iga, G., Lopez-Ortiz, C., Gracia-Rodriguez, C., Almeida, A., Nimmakayala, P., Reddy, U. K., & Balagurusamy, N. (2023). A Genome-Wide Identification and Comparative Analysis of the Heavy-Metal-Associated Gene Family in Cucurbitaceae Species and Their Role in Cucurbita pepo under Arsenic Stress. Genes, 14(10), 1877. https://doi.org/10.3390/genes14101877







