Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic
Abstract
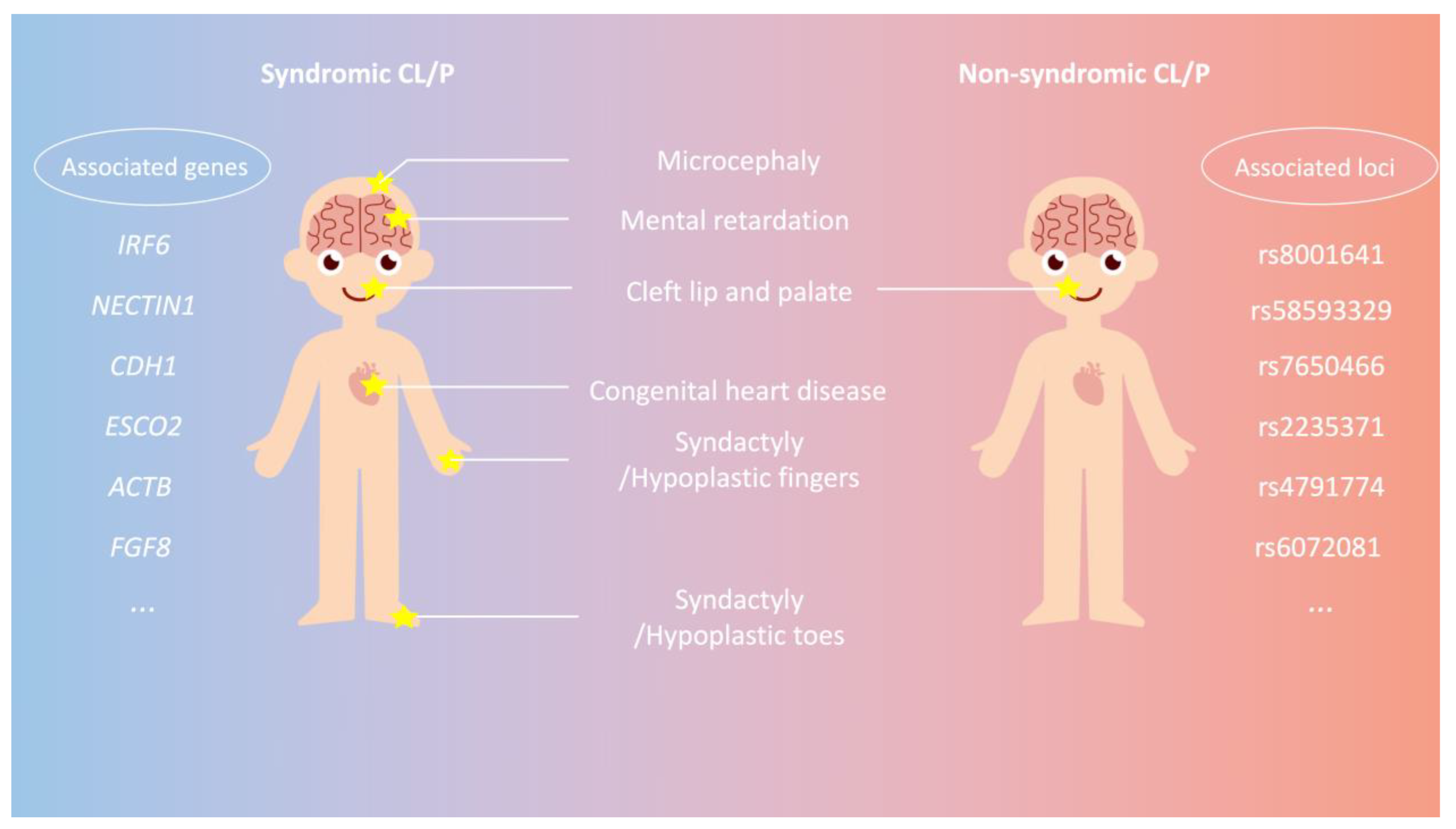
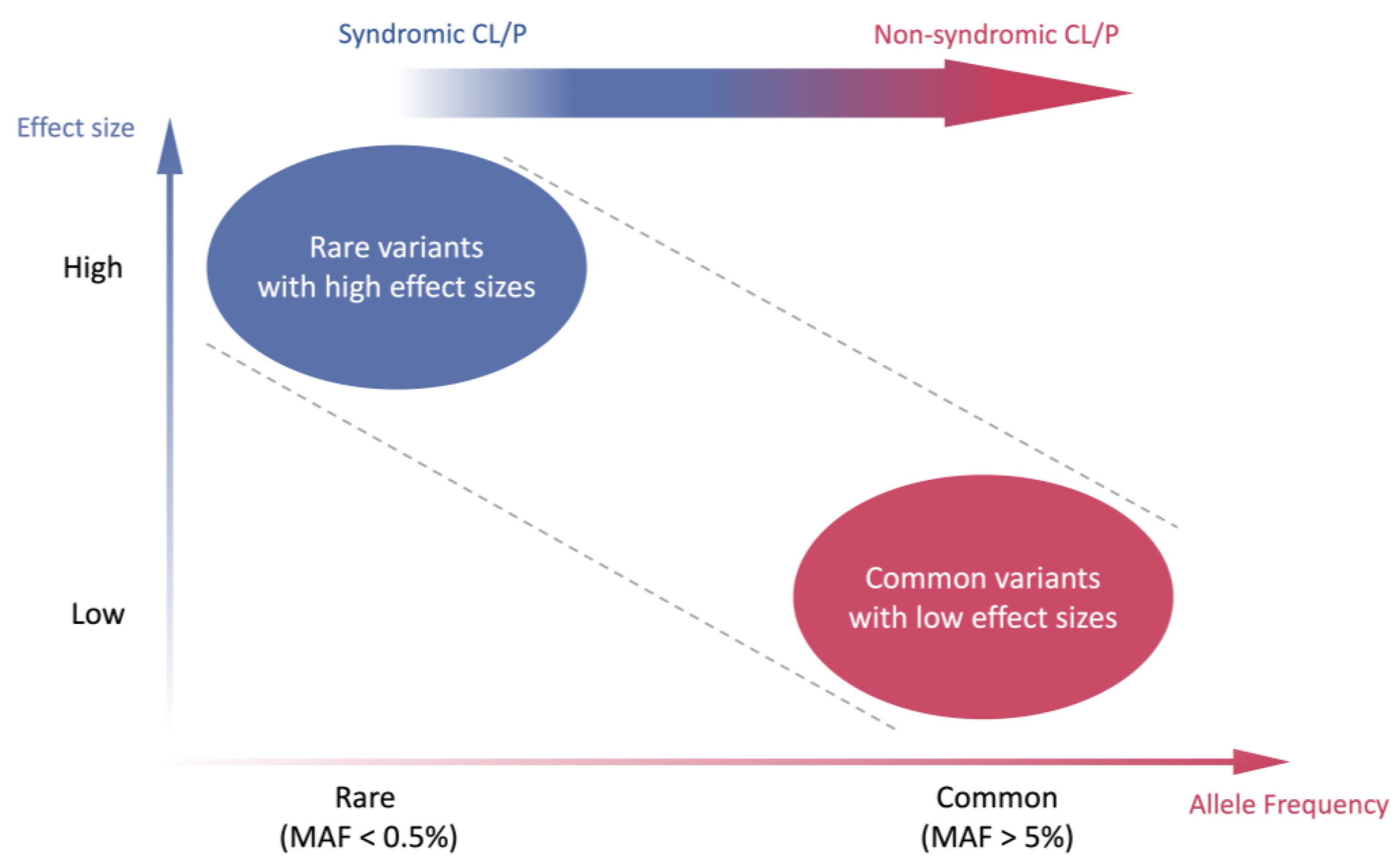
1. Background
2. Genetic Etiology of CL/P
| Gene | Location | Protein Function | Phenotype | Phenotype MIM Number † | Inheritance |
|---|---|---|---|---|---|
| ACTB | 7p22.1 | β Actin | Baraitser–Winter syndrome 1 | 243310 | AD |
| CDH1 | 16q22.1 | Cadherin 1 | Blepharocheilodontic syndrome 1 | 119580 | AD |
| EFNB1 | Xq13.1 | Ephrin B1 receptor protein-tyrosine kinase | Craniofrontonasal dysplasia | 304110 | XLD |
| ESCO2 | 8p21.1 | Chromatid cohesion N-acetyltransferase 2 | Juberg–Hayward syndrome | 216100 | AR |
| Roberts-SC phocomelia syndrome | 268300 | AR | |||
| FGF8 | 10q24.32 | Fibroblast Growth Factor 8 | Hypogonadotropic hypogonadism 6 with or without anosmia | 612702 | AD |
| GLI2 | 2q14.2 | GLI family zinc finger 2 | Holoprosencephaly 9 | 610829 | AD |
| GLI3 | 7p14.1 | GLI family zinc finger 3 | Pallister–Hall syndrome | 146510 | AD |
| HYLS1 | 11q24.2 | HYLS1 Centriolar and Ciliogenesis Associated | Hydrolethalus syndrome | 236680 | AR |
| IRF6 | 1q32.2 | Interferon regulatory 6 transcription factor | Van der Woude syndrome 1 | 119300 | AD |
| KDM6A | Xp11.3 | Lysine demethylase 6A | Kabuki syndrome 2 | 300867 | XLD |
| MID1 | Xp22.2 | Midline 1 | Opitz GBBB syndrome | 300000 | XLR |
| MSX1 | 4p16.2 | Msh homeobox 1 | Wolf–Hirschhorn syndrome | 194190 | Unknown [24,26] |
| NECTIN1 | 11q23.3 | Nectin cell adhesion molecule 1 | Cleft lip/palate-ectodermal dysplasia syndrome | 225060 | AR |
| OFD1 | Xp22.2 | Centriole and centriolar satellite protein | Orofaciodigital syndrome I | 311200 | XLD |
| PHF8 | Xp11.22 | PHD finger protein 8 | Intellectual developmental disorder, X-linked syndromic, Siderius type | 300263 | XLR |
| RIPK4 | 21q22.3 | Receptor interacting serine/threonine kinase 4 | Popliteal pterygium syndrome, Bartsocas–Papas type 1 | 263650 | AR |
| TFAP2A | 6p24.3 | Transcription factor AP-2 α | Branchiooculofacial syndrome | 113620 | AD |
| TP63 | 3q28 | Tumor protein p63 | Ectrodactyly, ectodermal dysplasia, and cleft lip/palate syndrome 3 | 604292 | AD |
| Hay–Wells syndrome | 106260 | AD | |||
| Rapp–Hodgkin syndrome | 129400 | AD | |||
| WNT3 | 17q21.31-q21.32 | Wnt family member 3 | Tetra-amelia syndrome 1 | 273395 | AR |
3. Genome-Wide Association Study (GWAS) of NSCL/P Worldwide
4. Association Studies and Related Loci of NSCL/P in the Chinese Population
| SNP ID | Affected Allele | Gene/Region | OR (95% CI) | p-Value | Population | Method | Case: Control | Year | PMID |
|---|---|---|---|---|---|---|---|---|---|
| rs7650466 | T | EPHA3 | 0.211(0.131–0.338) | 4.88 × 10−10 | Han Chinese | Targeted sequencing | 180:167 | 2018 | 29932736 [43] |
| rs58593329 | A | VAX1 | 1.34 (1.2–1.5) | 1.90 × 10−7 | Western Han Chinese | Targeted sequencing | 1626:2255 | 2022 | 35419918 [44] |
| rs11197887 | A | 1.35 (1.21–1.51) | 8.52 × 10−8 | ||||||
| rs1904302 | T | 1.39 (1.24–1.54) | 2.66 × 10−9 | ||||||
| rs10886040 | G | 1.40 (1.26–1.56 | 9.50 × 10−10 | ||||||
| rs744937 | T | 1.39 (1.25–1.54) | 2.23 × 10−9 | ||||||
| rs7078160 | A | 1.40 (1.25–1.55) | 1.12 × 10−9 | ||||||
| rs17095681 | T | SHTN1 | 0.64 (NA) | 3.80 × 10−9 | Han and Hui Chinese | GWAS and Targeted sequencing | 1931:2258 | 2016 | 28008912 [45] |
| rs4791331 | T | NTN1 | 1.43 (1.20–1.70) | 5.10 × 10−5 | Han Chinese | Targeted sequencing | 873:830 | 2020 | 31780810 [46] |
| rs2235371 | T | 1q32.2 | 0.67 (0.62–0.73) | 8.69 × 10−22 | Chinese | GWAS | 858:1248 | 2015 | 25775280 [47] |
| rs7078160 | A | 10q25.3 | 1.29 (1.19–1.39) | 3.09 × 10−10 | |||||
| rs8049367 | T | 16p13.3 | 0.74 (0.68–0.80) | 8.98 × 10−12 | |||||
| rs4791774 | G | 17p13.1 | 1.56 (1.42–1.72) | 5.05 × 10−19 | |||||
| rs13041247 | C | 20q12 | 0.76 (0.71–0.83) | 1.69 × 10−11 | |||||
| rs17820943 | T | 20q12 | - | 6.70 × 10−5 | Southern Han Chinese | Targeted sequencing | 430:451 | 2020 | 31713353 [48] |
| rs6072081 | G | - | 4.52 × 10−4 | ||||||
| rs6072081 | G | 20q12 | 0.72 (0.58–0.9) | 4.00 × 10−3 | Han Chinese | Targeted sequencing | 305:356 | 2012 | 22522387 [49] |
| rs13041247 | C | 0.68 (0.54–0.85) | 7.20 × 10−4 | ||||||
| rs6102085 | A | 0.62 (0.49–0.77) | 2.14 × 10−5 |
4.1. VAX1
4.2. 20q12
5. Polygenic Inheritance and Polygenic Risk Score in NSCL/P
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDH1 | Cadherin-1 |
| CL | cleft lip |
| CL/P | cleft lip with or without palate |
| CLPED1 | cleft lip/palate-ectodermal dysplasia syndrome |
| CNVs | copy number variants |
| CP | cleft palate |
| CTNND1 | Catenin Delta 1 |
| ESRP2 | Epithelial Splicing Regulatory Protein 2 |
| GWAS | genome-wide association study |
| IRF6 | Interferon Regulatory Factor 6 |
| MSX1 | Msh Homeobox 1 |
| NECTIN1 | Nectin Cell Adhesion Molecule 1 |
| NGS | next-generation sequencing |
| NSCL/P | non-syndromic cleft lip with or without palate |
| OFCs | orofacial clefts |
| PDGFRA | Platelet-Derived Growth Factor Receptor Alpha |
| PLEKHA5 | Pleckstrin Homology Domain-Containing Family A Member 5 |
| PRS | polygenic risk score |
| SCL/P | syndromic cleft lip with or without palate |
| SNPs | single nucleotide polymorphisms |
| SNVs | single nucleotide variants |
| VAX1 | Ventral Anterior Homeobox 1 |
| VWS | Van der Woude syndrome |
| WES | whole-exome sequencing |
| WGS | whole-genome sequencing |
References
- Rahimov, F.; Jugessur, A.; Murray, J.C. Genetics of Nonsyndromic Orofacial Clefts. Cleft Palate-Craniofac. J. 2012, 49, 73–91. [Google Scholar] [CrossRef]
- Nasreddine, G.; Hajj, J.E.; Ghassibe-Sabbagh, M. Orofacial clefts embryology, classification, epidemiology, and genetics. Mutat. Res. Mol. Mech. Mutagen. 2021, 787, 108373. [Google Scholar] [CrossRef]
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, N.; Georges, M.; Malevez, C.; Mey, A.D.; Mansbach, A. Embryology and epidemiology of cleft lip and palate. B-ENT 2006, 2, 11–19. [Google Scholar]
- Hammond, N.L.; Dixon, M.J. Author response for “Revisiting the embryogenesis of lip and palate development”. Oral Dis. 2022, 28, 1306–1326. [Google Scholar] [CrossRef]
- Murray, J.C. Gene/environment causes of cleft lip and/or palate. Clin. Genet. 2002, 61, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Chiquet, B.T.; Hashmi, S.S.; Henry, R.; Burt, A.; Mulliken, J.B.; Stal, S.; Bray, M.; Blanton, S.H.; Hecht, J.T. Genomic screening identifies novel linkages and provides further evidence for a role of MYH9 in nonsyndromic cleft lip and palate. Eur. J. Hum. Genet. 2009, 17, 195–204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tolarová, M.M.; Cervenka, J. Classification and birth prevalence of orofacial clefts. Am. J. Med. Genet. 1998, 75, 126–137. [Google Scholar] [CrossRef]
- Gorlin, R.J.; Cohen, M.M., Jr.; Hennekam, R.C. Syndromes of the Head and Neck; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Vanderas, A.P. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: A review. Cleft Palate J. 1987, 24, 216–225. [Google Scholar]
- Janečková, E.; Feng, J.; Li, J.; Rodriguez, G.; Chai, Y. Dynamic activation of WNT, FGF, and HH signaling during soft palate development. PLoS ONE 2019, 14, e0223879. [Google Scholar] [CrossRef]
- Won, H.-J.; Kim, J.-W.; Won, H.-S.; Shin, J.-O. Gene Regulatory Networks and Signaling Pathways in Palatogenesis and Cleft Palate: A Comprehensive Review. Cells 2023, 12, 1954. [Google Scholar] [CrossRef]
- Rizos, M.; Spyropoulos, M.N. Van der Woude syndrome: A review. Cardinal signs, epidemiology, associated features, differential diagnosis, expressivity, genetic counselling and treatment. Eur. J. Orthod. 2004, 26, 17–24. [Google Scholar] [CrossRef]
- Kondo, S.; Schutte, B.C.; Richardson, R.J.; Bjork, B.C.; Knight, A.S.; Watanabe, Y.; Howard, E.; de Lima, R.L.F.; Daack-Hirsch, S.; Sander, A.; et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002, 32, 285–289. [Google Scholar] [CrossRef]
- Yang, C.-W.; Yin, B.; Shi, J.-Y.; Shi, B.; Jia, Z.-L. Causal Variations at IRF6 Gene Identified in Van der Woude Syndrome Pedigrees. Cleft Palate Craniofac. J. 2023, 10556656231157575. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, X.; Xie, C.; Wu, B.; Song, G.; Du, Y. Detection of MSX1 gene mutations in patients with congenital tooth loss in Van der Woude syndrome. J. Prev. Treat. Stomatol. Dis. 2020, 28, 47–51. [Google Scholar] [CrossRef]
- Suzuki, K.; Hu, D.; Bustos, T.; Zlotogora, J.; Richieri-Costa, A.; Helms, J.A.; Spritz, R.A. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat. Genet. 2000, 25, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hayashi, R.; Fujita, H.; Kubota, M.; Kondo, M.; Shimomura, Y.; Niizeki, H. Novel homozygous mutation, c. 400C> T (p. Arg134*), in the PVRL 1 gene underlies cleft lip/palate-ectodermal dysplasia syndrome in an Asian patient. J. Dermatol. 2015, 42, 715–719. [Google Scholar] [CrossRef]
- Awadh, W.; Kiukkonen, A.; Nieminen, P.; Arte, S.; Hurmerinta, K.; Rice, D.P. Blepharocheilodontic (BCD) syndrome: New insights on craniofacial and dental features. Am. J. Med. Genet. Part A 2017, 173, 905–913. [Google Scholar] [CrossRef]
- Ghoumid, J.; Stichelbout, M.; Jourdain, A.-S.; Frenois, F.; Lejeune-Dumoulin, S.; Alex-Cordier, M.-P.; Lebrun, M.; Guerreschi, P.; Duquennoy-Martinot, V.; Vinchon, M.; et al. Blepharocheilodontic syndrome is a CDH1 pathway–related disorder due to mutations in CDH1 and CTNND1. Genet. Med. 2017, 19, 1013–1021. [Google Scholar] [CrossRef]
- Kievit, A.; Tessadori, F.; Douben, H.; Jordens, I.; Maurice, M.; Hoogeboom, J.; Hennekam, R.; Nampoothiri, S.; Kayserili, H.; Castori, M.; et al. Variants in members of the cadherin–catenin complex, CDH1 and CTNND1, cause blepharocheilodontic syndrome. Eur. J. Hum. Genet. 2018, 26, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Abu-Hussein, M.; Watted, N.; Yehia, M.; Proff, P.; Iraqi, F. Clinical genetic basis of tooth agenesis. J. Dent. Med. Sci. 2015, 14, 68–77. [Google Scholar]
- Liang, J.; Von den Hoff, J.; Lange, J.; Ren, Y.; Bian, Z.; Carels, C.E. MSX1 mutations and associated disease phenotypes: Genotype-phenotype relations. Eur. J. Hum. Genet. 2016, 24, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, P.; Kotilainen, J.; Aalto, Y.; Knuutila, S.; Pirinen, S.; Thesleff, I. MSX1 Gene is Deleted in Wolf-Hirschhorn Syndrome Patients with Oligodontia. J. Dent. Res. 2003, 82, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.; Hirschhorn, J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Rjiba, K.; Ayech, H.; Kraiem, O.; Slimani, W.; Jelloul, A.; Ben Hadj Hmida, I.; Mahdhaoui, N.; Saad, A.; Mougou-Zerelli, S. Disorders of sex development in Wolf–Hirschhorn syndrome: A genotype–phenotype correlation and MSX1 as candidate gene. Mol. Cytogenet. 2021, 14, 12. [Google Scholar] [CrossRef]
- Cox, L.L.; Cox, T.C.; Uribe, L.M.M.; Zhu, Y.; Richter, C.T.; Nidey, N.; Standley, J.M.; Deng, M.; Blue, E.; Chong, J.X.; et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018, 102, 1143–1157. [Google Scholar] [CrossRef]
- Ichikawa, E.; Watanabe, A.; Nakano, Y.; Akita, S.; Hirano, A.; Kinoshita, A.; Kondo, S.; Kishino, T.; Uchiyama, T.; Niikawa, N.; et al. PAX9 and TGFB3 are linked to susceptibility to nonsyndromic cleft lip with or without cleft palate in the Japanese: Population-based and family-based candidate gene analyses. J. Hum. Genet. 2006, 51, 38–46. [Google Scholar] [CrossRef]
- Lee, J.K.; Park, J.W.; Kim, Y.H.D.; Baek, S.-H.D. Association Between PAX9 Single-Nucleotide Polymorphisms and Nonsyndromic Cleft Lip With or Without Cleft Palate. J. Craniofac. Surg. 2012, 23, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Marazita, M.L. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. Part C Semin. Med. Genet. 2013, 163, 246–258. [Google Scholar] [CrossRef]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Mangold, E.; Herms, S.; Nowak, S.; Reutter, H.; Paul, A.; Becker, J.; Herberz, R.; AlChawa, T.; Nasser, E.; et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012, 44, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.L.; Chai, Y.; Yao, C.A.; Magee, W.; Figueiredo, J.C. Epidemiology, Etiology, and Treatment of Isolated Cleft Palate. Front. Physiol. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Leslie, E.J.; Cooper, M.E.; Butali, A.; Standley, J.; Rigdon, J.; Suzuki, S.; Gongorjav, A.; Shonkhuuz, T.E.; Natsume, N.; et al. Replication of 13q31.1 association in nonsyndromic cleft lip with cleft palate in Europeans. Am. J. Med. Genet. Part A 2015, 167, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Dunkhase, E.; Ludwig, K.U.; Knapp, M.; Skibola, C.F.; Figueiredo, J.C.; Hosking, F.J.; Ellinghaus, E.; Landi, M.T.; Ma, H.; Nakagawa, H.; et al. Nonsyndromic cleft lip with or without cleft palate and cancer: Evaluation of a possible common genetic background through the analysis of GWAS data. Genom. Data 2016, 10, 22–29. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Peterson, R.E.; Kuchenbaecker, K.; Walters, R.K.; Chen, C.-Y.; Popejoy, A.B.; Periyasamy, S.; Lam, M.; Iyegbe, C.; Strawbridge, R.J.; Brick, L.; et al. Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell 2019, 179, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Troubat, L.; Fettahoglu, D.; Aschard, H.; Julienne, H. Multi-trait GWAS for diverse ancestries: Mapping the knowledge gap. bioRxiv 2023. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Zhou, W.; Nielsen, J.B.; Fritsche, L.G.; Dey, R.; Gabrielsen, M.E.; Wolford, B.N.; LeFaive, J.; VandeHaar, P.; Gagliano, S.A.; Gifford, A.; et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018, 50, 1335–1341. [Google Scholar] [CrossRef]
- Atkinson, E.G.; Maihofer, A.X.; Kanai, M.; Martin, A.R.; Karczewski, K.J.; Santoro, M.L.; Ulirsch, J.C.; Kamatani, Y.; Okada, Y.; Finucane, H.K.; et al. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat. Genet. 2021, 53, 195–204. [Google Scholar] [CrossRef]
- Atkinson, E.G.; Bianchi, S.B.; Ye, G.Y.; Martínez-Magaña, J.J.; Tietz, G.E.; Montalvo-Ortiz, J.L.; Giusti-Rodriguez, P.; Palmer, A.A.; Sanchez-Roige, S. Cross-ancestry genomic research: Time to close the gap. Neuropsychopharmacology 2022, 47, 1737–1738. [Google Scholar] [CrossRef]
- Chen, R.; Guo, S.; Wang, X.; Mu, Y.; Duan, E.; Xu, Y. Association of EPHA3 Gene Polymorphisms with Nonsyndromic Cleft Lip With or Without Cleft Palate. Genet. Test. Mol. Biomark. 2018, 22, 420–424. [Google Scholar] [CrossRef]
- You, Y.; Shi, J.; Shi, B.; Jia, Z. Target sequencing reveals the association between variants in VAX1 and NSCL/P in Chinese population. Oral Dis. 2022, 29, 2130–2138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Huang, Y.; Pan, Y.; Yin, A.; Shi, B.; Du, X.; Ma, L.; Lan, F.; Jiang, M.; et al. Validation of a genome-wide association study implied that SHTIN1 may involve in the pathogenesis of NSCL/P in Chinese population. Sci. Rep. 2016, 6, 38872. [Google Scholar] [CrossRef]
- Li, D.; Zhu, G.; Lou, S.; Ma, L.; Zhang, C.; Pan, Y.; Wang, L. The functional variant of NTN1 contributes to the risk of nonsyndromic cleft lip with or without cleft palate. Eur. J. Hum. Genet. 2020, 28, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, Y.; Yin, A.; Pan, Y.; Wang, Y.; Wang, C.; Du, Y.; Wang, M.; Lan, F.; Hu, Z.; et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun. 2015, 6, 6414. [Google Scholar] [CrossRef]
- He, Y.; Huang, L.; Zheng, Y.; Chen, J.; Tang, S. Association of single nucleotide polymorphisms at 20q12 with nonsyndromic cleft lip with or without cleft palate in a Southern Chinese Han cohort. Mol. Genet. Genom. Med. 2020, 8, e1028. [Google Scholar] [CrossRef]
- Huang, E.; Cheng, H.; Xu, M.; Shu, S.; Tang, S. Association between single-nucleotide polymorphisms on chromosome 1p22 and 20q12 and nonsyndromic cleft lip with or without cleft palate: New data in Han Chinese and meta-analysis. Birth Defects Res. Part A Clin. Mol. Teratol. 2012, 94, 469–476. [Google Scholar] [CrossRef]
- Geoghegan, F.; Xavier, G.; Birjandi, A.; Seppala, M.; Cobourne, M. Vax1 Plays an Indirect Role in the Etiology of Murine Cleft Palate. J. Dent. Res. 2017, 96, 1555–1562. [Google Scholar] [CrossRef]
- Beaty, T.H.; Murray, J.C.; Marazita, M.L.; Munger, R.G.; Ruczinski, I.; Hetmanski, J.B.; Liang, K.Y.; Wu, T.; Murray, T.; Fallin, M.D.; et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010, 42, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-H.; Shi, J.-Y.; Lin, Y.-S.; Shi, B.; Jia, Z.-L. VAX1 gene associated non-syndromic cleft lip with or without palate in Western Han Chinese. Arch. Oral Biol. 2018, 95, 40–43. [Google Scholar] [CrossRef]
- Beaty, T.H.; Taub, M.A.; Scott, A.F.; Murray, J.C.; Marazita, M.L.; Schwender, H.; Parker, M.; Hetmanski, J.B.; Balakrishnan, P.; Mansilla, M.A.; et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case–parent trio study. Hum Genet. 2013, 132, 771–781. [Google Scholar] [CrossRef]
- Gurramkonda, V.B.; Syed, A.H.; Murthy, J.; Chaubey, G.; Lakkakula, V.B. Polymorphic variants near 1p22 and 20q11.2 loci and the risk of non-syndromic cleft lip and palate in South Indian population. Int. J. Pediatr. Otorhinolaryngol. 2015, 79, 2389–2393. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Mak, T.S.-H.; O’reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lee, L.; Chen, J.; Collins, R.; Wu, F.; Guo, Y.; Linksted, P.; Peto, R. Cohort Profile: The Kadoorie Study of Chronic Disease in China (KSCDC). Int. J. Epidemiol. 2005, 34, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Wade, K.H.; Zahid, S.; Brancale, J.; Xia, R.; Distefano, M.; Senol-Cosar, O.; Haas, M.E.; Bick, A.; et al. Polygenic Prediction of Weight and Obesity Trajectories from Birth to Adulthood. Cell 2019, 177, 587–596.e9. [Google Scholar] [CrossRef]
- Dai, J.; Lv, J.; Zhu, M.; Wang, Y.; Qin, N.; Ma, H.; He, Y.-Q.; Zhang, R.; Tan, W.; Fan, J.; et al. Identification of risk loci and a polygenic risk score for lung cancer: A large-scale prospective cohort study in Chinese populations. Lancet Respir. Med. 2019, 7, 881–891. [Google Scholar] [CrossRef]
- Vilhjálmsson, B.J.; Yang, J.; Finucane, H.K.; Gusev, A.; Lindström, S.; Ripke, S.; Genovese, G.; Loh, P.-R.; Bhatia, G.; Do, R.; et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet. 2015, 97, 576–592. [Google Scholar] [CrossRef]
- Lloyd-Jones, L.R.; Zeng, J.; Sidorenko, J.; Yengo, L.; Moser, G.; Kemper, K.E.; Wang, H.; Zheng, Z.; Magi, R.; Esko, T.; et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019, 10, 5086. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Chen, C.-Y.; Ni, Y.; Feng, Y.-C.A.; Smoller, J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience 2019, 8, giz082. [Google Scholar] [CrossRef] [PubMed]
- Kramer, I.; Hooning, M.J.; Mavaddat, N.; Hauptmann, M.; Keeman, R.; Steyerberg, E.W.; Giardiello, D.; Antoniou, A.C.; Pharoah, P.D.; Canisius, S.; et al. Breast Cancer Polygenic Risk Score and Contralateral Breast Cancer Risk. Am. J. Hum. Genet. 2020, 107, 837–848. [Google Scholar] [CrossRef]
- Mars, N.; Widén, E.; Kerminen, S.; Meretoja, T.; Pirinen, M.; Parolo, P.d.B.; Palta, P.; Havulinna, A.; Elliott, A.; Shcherban, A.; et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat. Commun. 2020, 11, 6383. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Z.; Cui, Q.; Liu, F.; Li, J.; Niu, X.; Shen, C.; Hu, D.; Huang, K.; Chen, J.; et al. A polygenic risk score improves risk stratification of coronary artery disease: A large-scale prospective Chinese cohort study. Eur. Hear. J. 2022, 43, 1702–1711. [Google Scholar] [CrossRef]
- Koyama, S.; Ito, K.; Terao, C.; Akiyama, M.; Horikoshi, M.; Momozawa, Y.; Matsunaga, H.; Ieki, H.; Ozaki, K.; Onouchi, Y.; et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020, 52, 1169–1177. [Google Scholar] [CrossRef]
- Howe, L.J.; Lee, M.K.; Sharp, G.C.; Smith, G.D.; Pourcain, B.S.; Shaffer, J.R.; Ludwig, K.U.; Mangold, E.; Marazita, M.L.; Feingold, E.; et al. Investigating the shared genetics of non-syndromic cleft lip/palate and facial morphology. PLoS Genet. 2018, 14, e1007501. [Google Scholar] [CrossRef]
- Blanco, R.; Colombo, A.; Suazo, J. Genetic risk score for nonsyndromic cleft lip with or without cleft palate for a Chilean population. Genet. Couns. 2014, 25, 143–149. [Google Scholar]
- Yu, Y.; Alvarado, R.; Petty, L.E.; Bohlender, R.J.; Shaw, D.M.; Below, J.E.; Bejar, N.; Ruiz, O.E.; Tandon, B.; Eisenhoffer, G.T.; et al. Polygenic risk impacts PDGFRA mutation penetrance in non-syndromic cleft lip and palate. Hum. Mol. Genet. 2022, 31, 2348–2357. [Google Scholar] [CrossRef]
- Fell, M.; Dack, K.; Chummun, S.; Sandy, J.; Wren, Y.; Lewis, S. Maternal Cigarette Smoking and Cleft Lip and Palate: A Systematic Review and Meta-Analysis. Cleft Palate-Craniofac. J. 2022, 59, 1185–1200. [Google Scholar] [CrossRef]
- Kurita, H.; Motoki, N.; Inaba, Y.; Misawa, Y.; Ohira, S.; Kanai, M.; Tsukahara, T.; Nomiyama, T.; Kamijima, M.; Yamazaki, S.; et al. Maternal alcohol consumption and risk of offspring with congenital malformation: The Japan Environment and Children’s Study. Pediatr. Res. 2021, 90, 479–486. [Google Scholar] [CrossRef]
- Proctor-Williams, K.; Louw, B. Cleft Lip and/or Palate in Infants Prenatally Exposed to Opioids. Cleft Palate-Craniofac. J. 2022, 59, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Alrbata, R.; Almaaiteh, H.; Albdour, M.; Alshammout, R. A retrospective cohort study to evaluate the association between types of nonsyndromic oral clefts and a child’s gender and maternal age. J. Int. Soc. Prev. Community Dent. 2021, 11, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-P.; Qu, W.-D.; Sun, C.; Cao, R.-Y.; Liu, D.-W.; Du, P.-G. A study on environmental factors for nonsyndromic cleft lip and/or palate. J. Craniofac. Surg. 2018, 29, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Qi, Y.P.; Yeung, L.F.; Mai, C.T.; Zauche, L.H.; Wang, A.; Daniels, K.; Williams, J.L. Folic Acid and the Prevention of Birth Defects: 30 Years of Opportunity and Controversies. Annu. Rev. Nutr. 2022, 42, 423–452. [Google Scholar] [CrossRef]
- Lin, Y.; Shu, S.; Tang, S. A case-control study of environmental exposures for nonsyndromic cleft of the lip and/or palate in eastern Guangdong, China. Int. J. Pediatr. Otorhinolaryngol. 2014, 78, 545–551. [Google Scholar] [CrossRef]
- Mahapure, K.S.; Powar, R.S. Could maternal stress be a causal factor for nonsyndromic cleft lip and/or palate: A retrospective study. Natl. J. Maxillofac. Surg. 2022, 13, S36–S40. [Google Scholar] [CrossRef]
- Johnsen, P.V.; Riemer-Sørensen, S.; DeWan, A.T.; Cahill, M.E.; Langaas, M. A new method for exploring gene–gene and gene–environment interactions in GWAS with tree ensemble methods and SHAP values. BMC Bioinform. 2021, 22, 230. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, X.; Du, F.; Long, X.; Huang, J. Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic. Genes 2023, 14, 1859. https://doi.org/10.3390/genes14101859
Cheng X, Du F, Long X, Huang J. Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic. Genes. 2023; 14(10):1859. https://doi.org/10.3390/genes14101859
Chicago/Turabian StyleCheng, Xi, Fengzhou Du, Xiao Long, and Jiuzuo Huang. 2023. "Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic" Genes 14, no. 10: 1859. https://doi.org/10.3390/genes14101859
APA StyleCheng, X., Du, F., Long, X., & Huang, J. (2023). Genetic Inheritance Models of Non-Syndromic Cleft Lip with or without Palate: From Monogenic to Polygenic. Genes, 14(10), 1859. https://doi.org/10.3390/genes14101859






