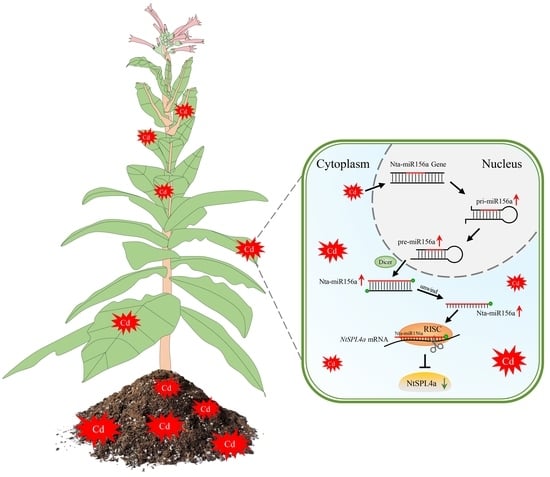
The NtSPL Gene Family in Nicotiana tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of the NtSPL and Nta-miR156 Gene Family
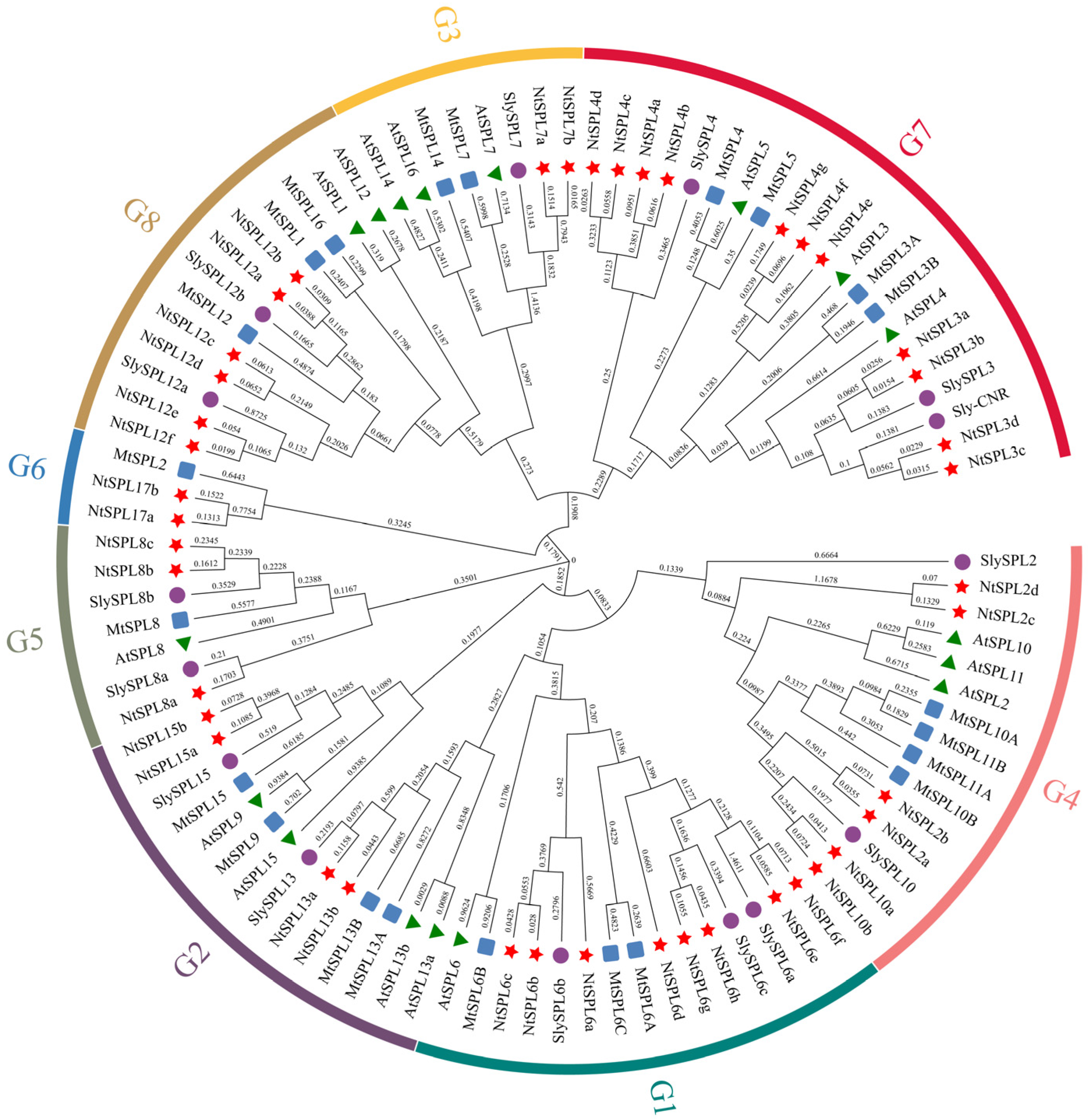
2.2. Phylogenetic Analysis
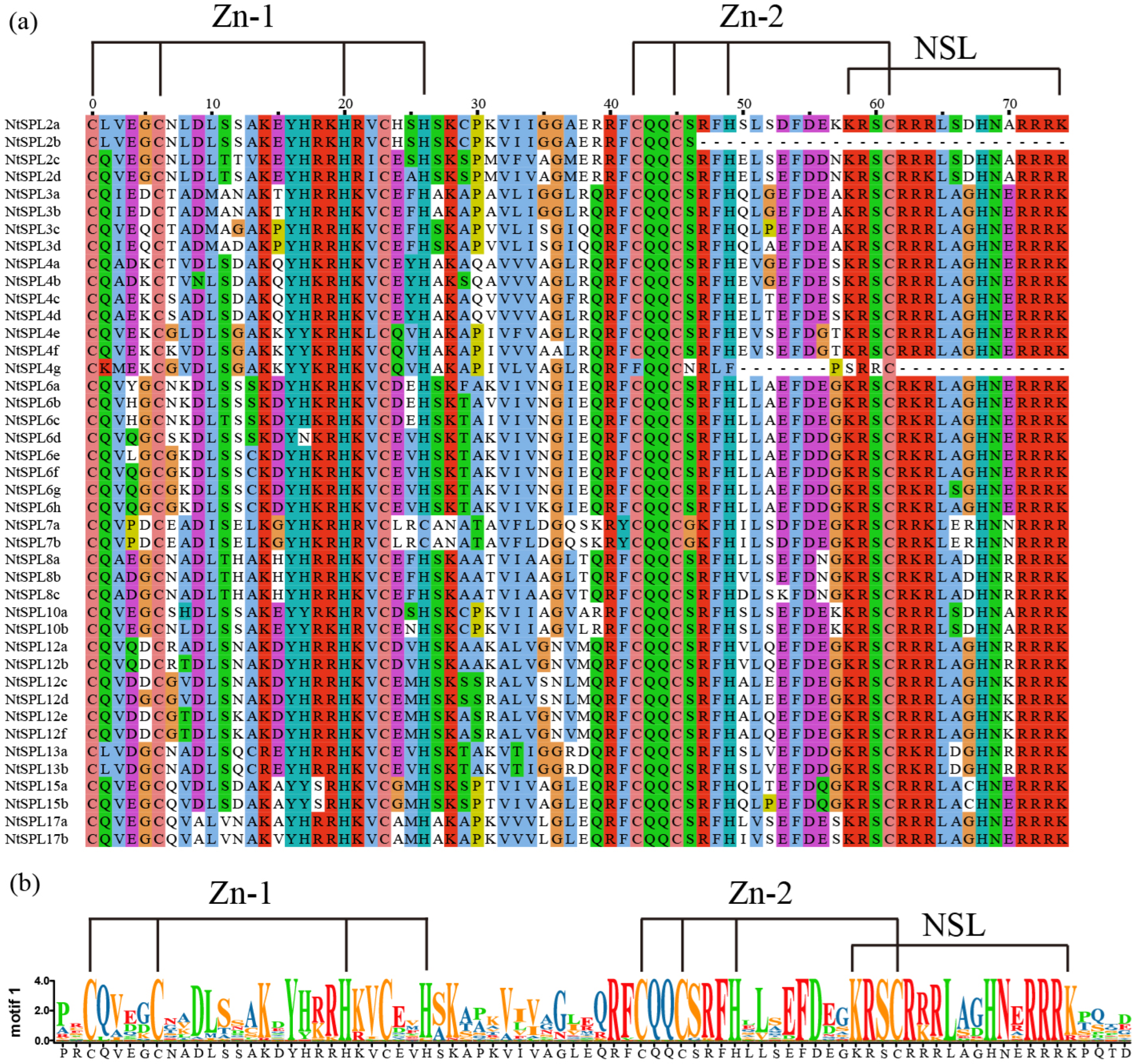
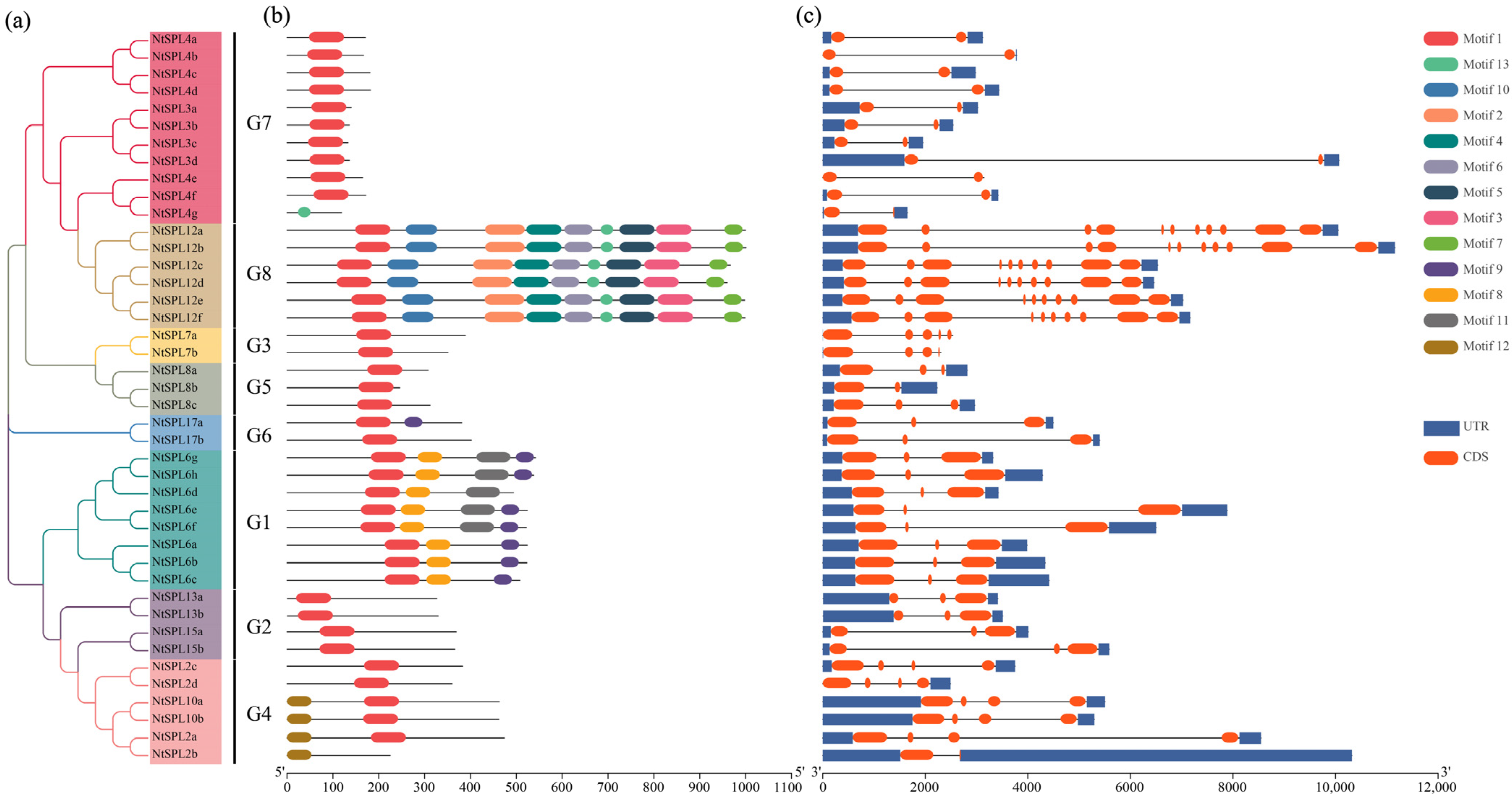
2.3. Multiple Sequence Alignment Analysis and Motif Composition and Gene Structure
2.4. Prediction of Cis-Acting Elements
2.5. Predictive Analysis of NtSPL Gene Family miRNAs
2.6. NtSPL Expression Patterns by Transcriptome Data
2.7. Plant Materials and Heavy Metal Treatments
2.8. Analysis of the Pattern of NtSPL Gene Expression
3. Results
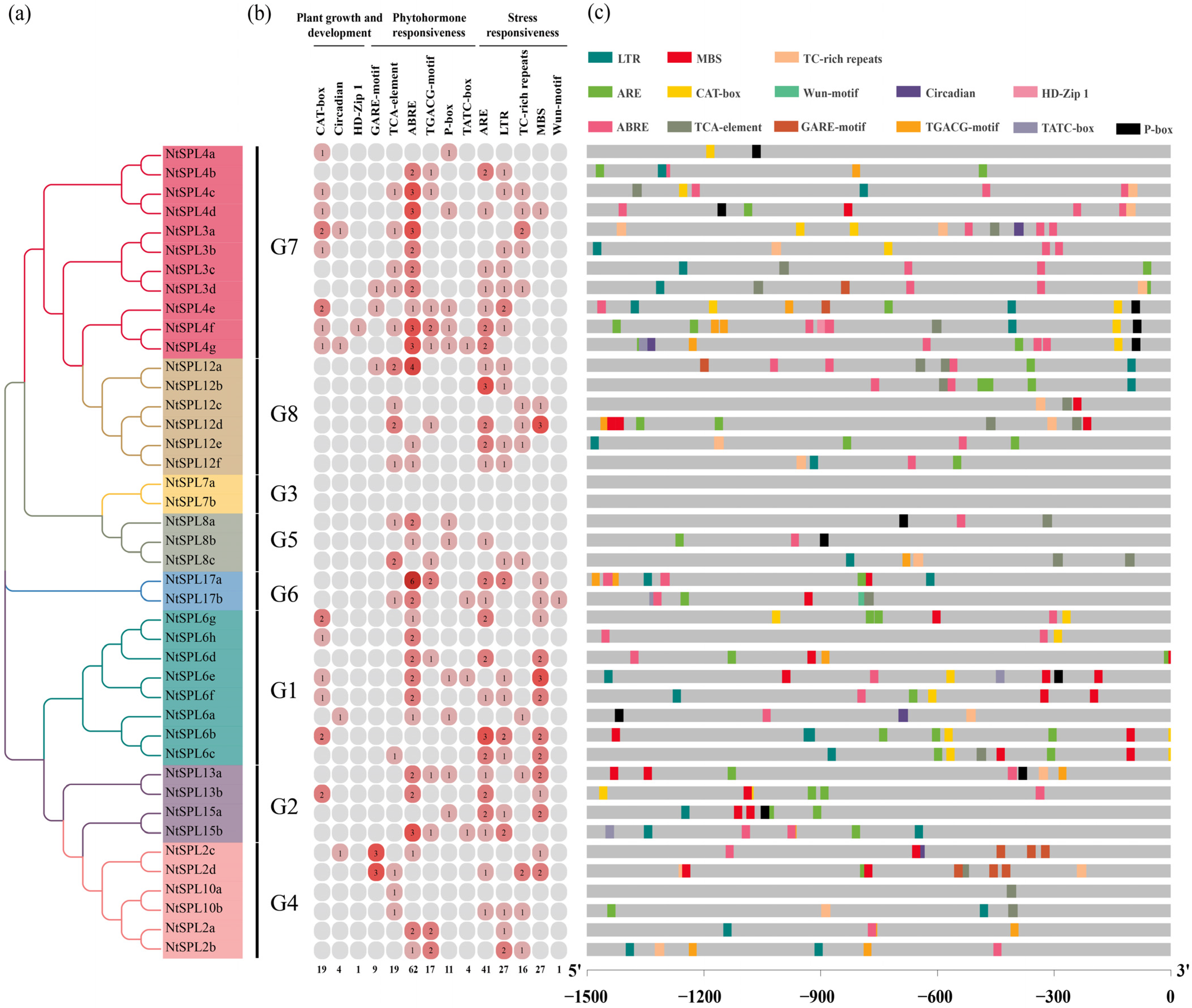
3.1. Identification and Phylogenetic Analysis of NtSPL Genes
3.2. Multiple Sequence Alignment of NtSPL Proteins
3.3. Conserved Motifs and Gene Structure Analysis of NtSPL Genes
3.4. Cis-Acting Elements in the Promoter Regions of NtSPL Genes
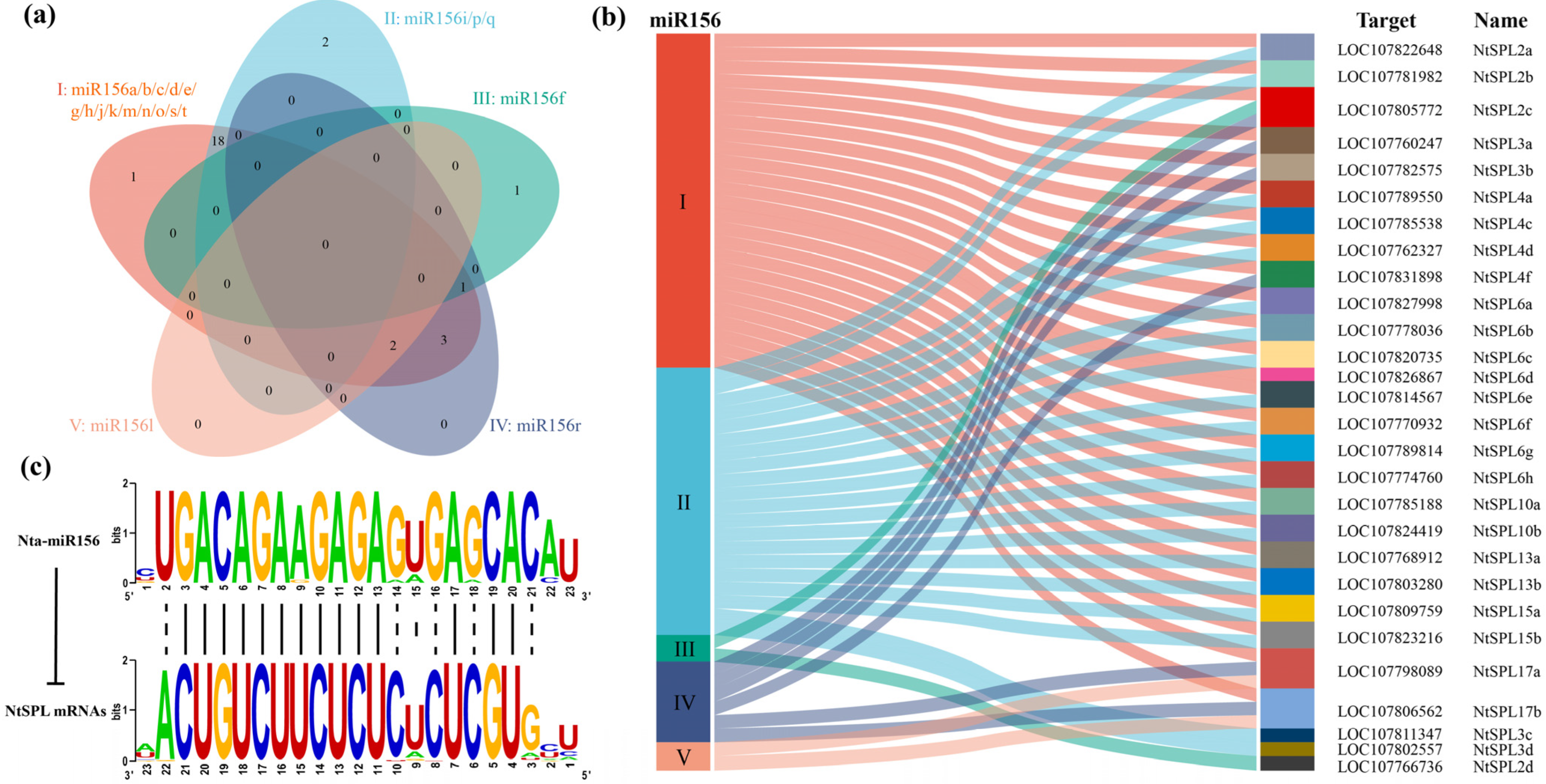
3.5. Nta-miR156 Multiplex Sequence Alignment
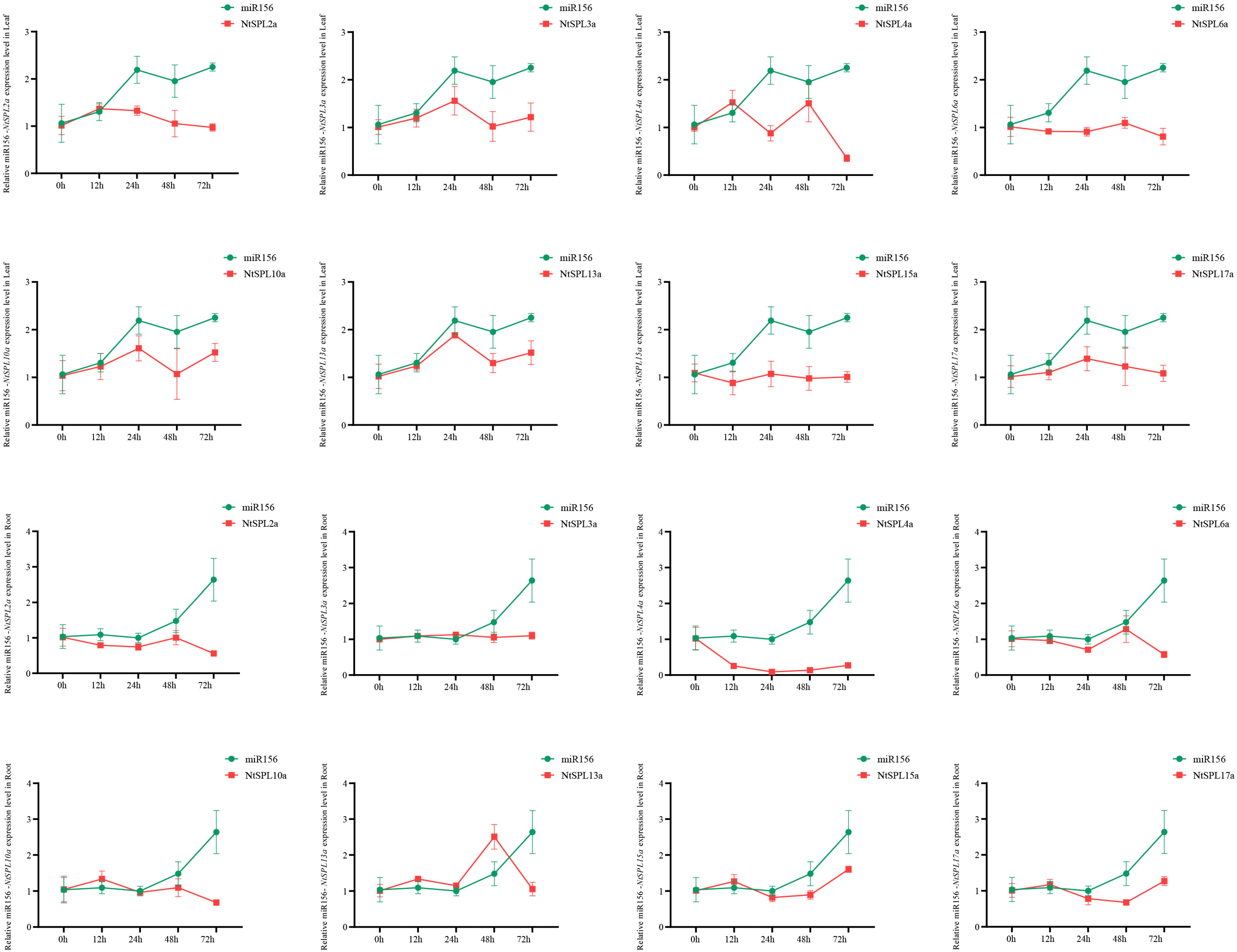
3.6. Analysis of miR156 and Its Target Sequences
3.7. Expression Pattern Analysis Based on Transcriptome Data
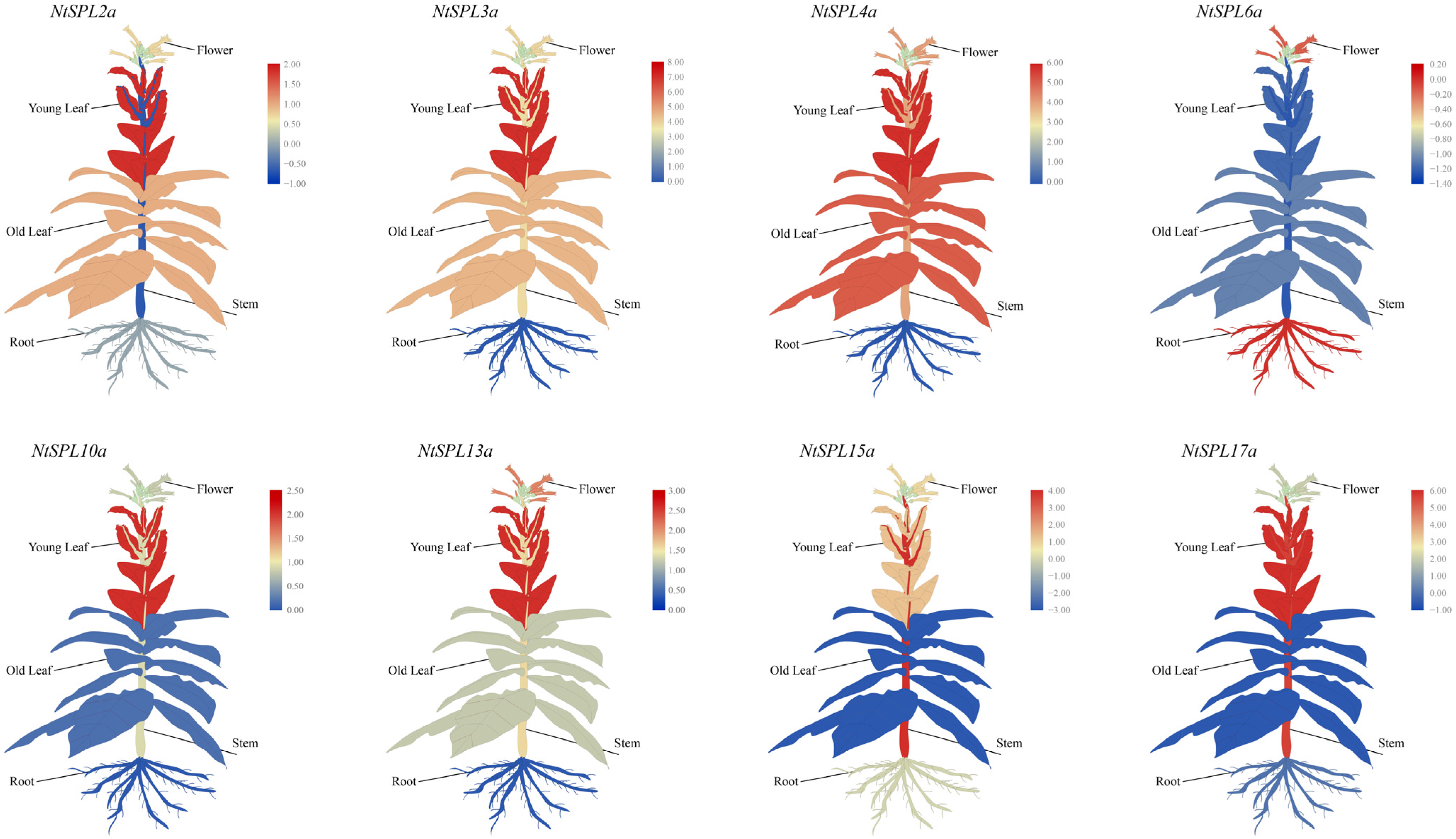
3.8. Tissue-Specific Expression Profiles of NtSPL Family Genes
3.9. Heavy Metal Stress-Induced Expression Profiles of NtSPL Family Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Li, Y.; Zhu, X.; Gui, X.; Ma, C.; Peng, W.; Li, Y.; Zhang, Y.; Huang, W.; Hua, D.; et al. Evaluation of the cadmium phytoextraction potential of tobacco (Nicotiana Tabacum) and rhizosphere micro-characteristics under different cadmium levels. Chemosphere 2022, 286, 131714. [Google Scholar] [CrossRef]
- Rehman, M.Z.U.; Rizwan, M.; Sohail, M.I.; Ali, S.; Waris, A.A.; Khalid, H.; Naeem, A.; Ahmad, H.R.; Rauf, A. Opportunities and challenges in the remediation of metal-contaminated soils by using tobacco (Nicotiana tabacum L.): A critical review. Environ. Sci. Pollut. Res. 2019, 26, 18053–18070. [Google Scholar] [CrossRef]
- Janaydeh, M.; Ismail, A.; Zulkifli, S.Z.; Omar, H. Toxic heavy metal (Pb and Cd) content in tobacco cigarette brands in selangor state, Peninsular Malaysia. Environ. Monit. Assess. 2019, 191, 637. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, D.J. Smoking, cadmium, and emphysema. Thorax 2004, 59, 184–185. [Google Scholar] [CrossRef]
- Lee, J.-W.; Kim, Y.; Kim, Y.; Yoo, H.; Kang, H.-T. Cigarette Smoking in Men and Women and Electronic Cigarette Smoking in Men are Associated with Higher Risk of Elevated Cadmium Level in the Blood. J. Korean Med. Sci. 2020, 35, e15. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA Promoter-Binding Protein-Like Transcription Factors: Star Players for Plant Growth and Development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, G.; Li, Y.; Yang, Z.; Liu, T.; Xie, X.; Kong, X.; Sun, J. PIL transcription factors directly interact with SPLs and repress tillering/branching in plants. New Phytol. 2022, 233, 1414–1425. [Google Scholar] [CrossRef]
- He, Y.; Fu, X.; Li, L.; Sun, X.; Tang, K.; Zhao, J. AaSPL9 affects glandular trichomes initiation by positively regulating expression of AaHD1 in Artemisia annua L. Plant Sci. 2022, 317, 111172. [Google Scholar] [CrossRef]
- Hyde, P.T.; Setter, T.L. Long-day photoperiod and cool temperature induce flowering in cassava: Expression of signaling genes. Front. Plant Sci. 2022, 13, 973206. [Google Scholar] [CrossRef]
- He, F.; Long, R.; Wei, C.; Zhang, Y.; Li, M.; Kang, J.; Yang, Q.; Wang, Z.; Chen, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family and its important role in salt stress in Medicago sativa L. BMC Plant Biol. 2022, 22, 295. [Google Scholar] [CrossRef]
- Chao, L.-M.; Liu, Y.-Q.; Chen, D.-Y.; Xue, X.-Y.; Mao, Y.-B.; Chen, X.-Y. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Z.; Li, Z.; Dong, S.; Yu, X.; Zhao, P.; Liao, W.; Yu, X.; Peng, M. MeSPL9 Attenuates Drought Resistance by Regulating JA Signaling and Protectant Metabolite Contents in Cassava. Theor. Appl. Genet. 2022, 135, 817–832. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, M.; Yu, J.; Guo, C. SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in Arabidopsis thaliana. BMC Plant Biol. 2022, 22, 59. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.; Ma, Y.; Wang, H.; Shi, Y. Role of transpiration and metabolism in translocation and accumulation of cadmium in tobacco plants (Nicotiana tabacum L.). Chemosphere 2016, 144, 1960–1965. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, J.; Jiang, D.; Hong, Y.; Xu, J.; Zheng, S.; Yang, J.; Chen, W. The miR157-SPL-CNR module acts upstream of bHLH101 to negatively regulate iron deficiency responses in tomato. JIPB 2022, 64, 1059–1075. [Google Scholar] [CrossRef]
- Mermod, M.; Takusagawa, M.; Kurata, T.; Kamiya, T.; Fujiwara, T.; Shikanai, T. SQUAMOSA promoter-binding protein-like 7 mediates copper deficiency response in the presence of high nitrogen in Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 835–846. [Google Scholar] [CrossRef]
- Garcia-Molina, A.; Xing, S.; Huijser, P. Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response. BMC Plant Biol. 2014, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Rong, F.; Liu, Z.; Ruan, S.; Lu, T.; Qian, H. Cadmium induced a non-Coding RNA microRNA535 mediates Cd accumulation in rice. J. Environ. Sci. 2022, 130, 149–162. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging Roles of MicroRNAs in Plant Heavy Metal Tolerance and Homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, S.; Deng, Y.; You, C.; Zhang, W.; Zhou, J.; Chen, X.; Gao, L.; Tang, Y. Genome-Wide mRNA and small RNA transcriptome profiles uncover cultivar- and tissue-specific changes induced by cadmium in Brassica Parachinensis. Environ. Exp. Bot. 2020, 180, 104207. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, H.; Jiang, H.; Wang, H.; Chen, K.; Duan, J.; Feng, S.; Wu, G. Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis. Chemosphere 2020, 242, 125168. [Google Scholar] [CrossRef]
- Teng, R.-M.; Wang, Y.-X.; Li, H.; Liu, H.; Wang, Y.; Zhuang, J. Identification and Expression Analysis of the SBP-box Gene Family Related to Abiotic Stress in Tea Plant (Camellia Sinensis (L.) O. Kuntze). Plant Mol. Biol. Rep. 2022, 40, 148–162. [Google Scholar] [CrossRef]
- Ma, L.; Liu, X.; Liu, W.; Wen, H.; Zhang, Y.; Pang, Y.; Wang, X. Characterization of Squamosa-Promoter Binding Protein-Box Family Genes Reveals the Critical Role of MsSPL20 in Alfalfa Flowering Time Regulation. Front. Plant Sci. 2021, 12, 775690. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Fan, Y.; Xue, G.; He, A.; Yang, H.; He, C.; Li, Y.; Ruan, J.; Yan, J.; Cheng, J. Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 389. [Google Scholar] [CrossRef]
- He, B.; Gao, S.; Lu, H.; Yan, J.; Li, C.; Ma, M.; Wang, X.; Chen, X.; Zhan, Y.; Zeng, F. Genome-wide analysis and molecular dissection of the SPL gene family in Fraxinus Mandshurica. BMC Plant Biol. 2022, 22, 451. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Lu, Q.; Wilson, I.W.; Qiu, D. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef]
- Zhao, H.; Cao, H.; Zhang, M.; Deng, S.; Li, T.; Xing, S. Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa. Genes 2022, 13, 1455. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, Y.; Zhang, N.; Zhang, X.; Wu, J.; Huang, Y.; Ruan, M.; Zhang, J.; Qi, Y. Systematic Identification, Evolution and Expression Analysis of the SPL Gene Family in Sugarcane (Saccharum spontaneum). Trop. Plant Biol. 2021, 14, 313–328. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-MPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From Data Annotation to Functional Exploration of Plant MicroRNAs. Nucleic Acids Res. 2022, 50, D1475–D1482. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Z.; Xu, Y.; Kong, L.; Shi, J.; Liu, Y.; Fu, C.; Wang, X.; Wang, Z.-Y.; Zhou, C.; et al. Genome-Wide characterization of SPL family in Medicago truncatula reveals the novel roles of miR156/SPL module in spiky pod development. BMC Genom. 2019, 20, 552. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. PsRNATarget: A Plant Small RNA Target Analysis Server (2017 Release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A Comprehensive, Interaction-Friendly Clinical Bioinformatics Analysis Platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The Tobacco Genome Sequence and Its Comparison with Those of Tomato and Potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Huo, C.; He, L.; Yu, T.; Ji, X.; Li, R.; Zhu, S.; Zhang, F.; Xie, H.; Liu, W. The Superoxide Dismutase Gene Family in Nicotiana tabacum: Genome-Wide Identification, Characterization, Expression Profiling and Functional Analysis in Response to Heavy Metal Stress. Front. Plant Sci. 2022, 13, 904105. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, J.; Li, Z.; Zhang, S.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. MiR156/157 Targets SPLs to Regulate Flowering Transition, Plant Architecture and Flower Organ Size in Petunia. Plant. Cell Physiol. 2021, 62, 839–857. [Google Scholar] [CrossRef]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in Soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Q.; Wang, J.-G.; Wang, L.-Y.; Wang, J.-F.; Wang, Q.; Yu, P.; Bai, M.-Y.; Fan, M. Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J. Integr. Plant Biol. 2020, 62, 421–432. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in Poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Y.; Liu, Y.; Peng, B.; Zhang, L.; Tang, K.; Chen, W. The SPB-Box Transcription Factor AaSPL2 Positively Regulates Artemisinin Biosynthesis in Artemisia annua L. Front. Plant. Sci. 2019, 10, 409. [Google Scholar] [CrossRef] [PubMed]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depège, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-G.; Shan, J.-X.; Shi, M.; Gao, J.-P.; Lin, H.-X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic Organization, Phylogenetic Comparison and Differential Expression of the SBP-Box Family Genes in Grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef] [PubMed]
- Shalom, L.; Shlizerman, L.; Zur, N.; Doron-Faigenboim, A.; Blumwald, E.; Sadka, A. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 2015, 6, 389. [Google Scholar] [CrossRef]
- Eriksson, M.; Moseley, J.L.; Tottey, S.; del Campo, J.A.; Quinn, J.; Kim, Y.; Merchant, S. Genetic Dissection of Nutritional Copper Signaling in Chlamydomonas Distinguishes Regulatory and Target Genes. Genetics 2004, 168, 795–807. [Google Scholar] [CrossRef]
- Guo, A.-Y.; Zhu, Q.-H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum Tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × Domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef]
- Mao, H.-D.; Yu, L.-J.; Li, Z.-J.; Yan, Y.; Han, R.; Liu, H.; Ma, M. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein–Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. Toxicity responses of Cu and Cd: The involvement of miRNAs and the transcription factor SPL7. BMC Plant Biol. 2016, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef]
- Wang, P.; Chen, D.; Zheng, Y.; Jin, S.; Yang, J.; Ye, N. Identification and expression analyses of SBP-Box Genes Reveal Their Involvement in Abiotic Stress and Hormone Response in Tea Plant (Camellia Sinensis). Int. J. Mol. Sci. 2018, 19, 3404. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yan, S.; Jing, Y.; Yang, R.; Zhang, Y.; Zhou, Y.; Zhu, Y.; Sun, J. MIR156-Targeted SPL9 Is Phosphorylated by SnRK2s and Interacts With ABI5 to Enhance ABA Responses in Arabidopsis. Front. Plant Sci. 2021, 12, 70853. [Google Scholar] [CrossRef]
- Lambertz, C.; Hemschemeier, A.; Happe, T. Anaerobic Expression of the Ferredoxin-Encoding FDX5 Gene of Chlamydomonas reinhardtii Is Regulated by the Crr1 Transcription Factor. Eukaryot. Cell 2010, 9, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Q.; Zhu, X.; Cui, M.; Jia, H.; Zhang, W.; Tang, W.; Leng, X.; Shen, W. Characterization on the Conservation and Diversification of MiRNA156 Gene Family from Lower to Higher Plant Species Based on Phylogenetic Analysis at the Whole Genomic Level. Funct. Integr. Genom. 2019, 19, 933–952. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.K.; Bregitzer, P.; Singh, J. Genome-Wide Analysis of the SPL/MiR156 Module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef]
- Li, X.-Y.; Lin, E.-P.; Huang, H.-H.; Niu, M.-Y.; Tong, Z.-K.; Zhang, J.-H. Molecular Characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family in Betula luminifera. Front. Plant Sci. 2018, 9, 608. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Peng, X.; Cao, H.; Yang, K.; Xiang, L.; Li, R.; Zhang, F.; Liu, W. The NtSPL Gene Family in Nicotiana tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress. Genes 2023, 14, 183. https://doi.org/10.3390/genes14010183
He L, Peng X, Cao H, Yang K, Xiang L, Li R, Zhang F, Liu W. The NtSPL Gene Family in Nicotiana tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress. Genes. 2023; 14(1):183. https://doi.org/10.3390/genes14010183
Chicago/Turabian StyleHe, Linshen, Xiang Peng, Hanping Cao, Kunjian Yang, Lien Xiang, Rui Li, Fangyuan Zhang, and Wanhong Liu. 2023. "The NtSPL Gene Family in Nicotiana tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress" Genes 14, no. 1: 183. https://doi.org/10.3390/genes14010183
APA StyleHe, L., Peng, X., Cao, H., Yang, K., Xiang, L., Li, R., Zhang, F., & Liu, W. (2023). The NtSPL Gene Family in Nicotiana tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress. Genes, 14(1), 183. https://doi.org/10.3390/genes14010183