Report of Hermansky–Pudlak Syndrome in Two Families with Novel Variants in HPS3 and HPS4 Genes
Abstract
1. Introduction
2. Material and Methods
2.1. Samples
2.2. Genomic DNA Extraction and Genetic Analysis
3. Results
3.1. Clinical Features of the Patients
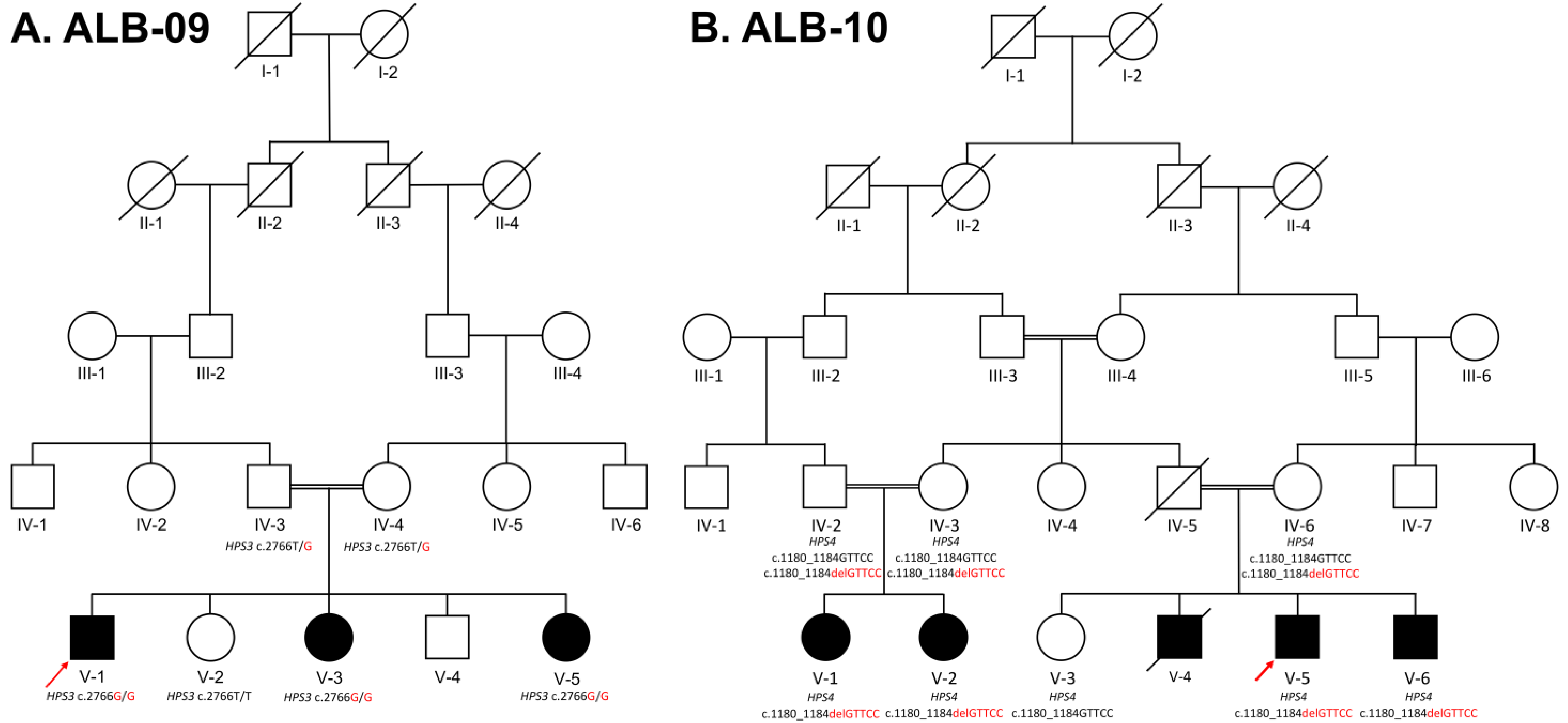
3.1.1. ALB-09 HPS3
3.1.2. ALB-10 HPS4
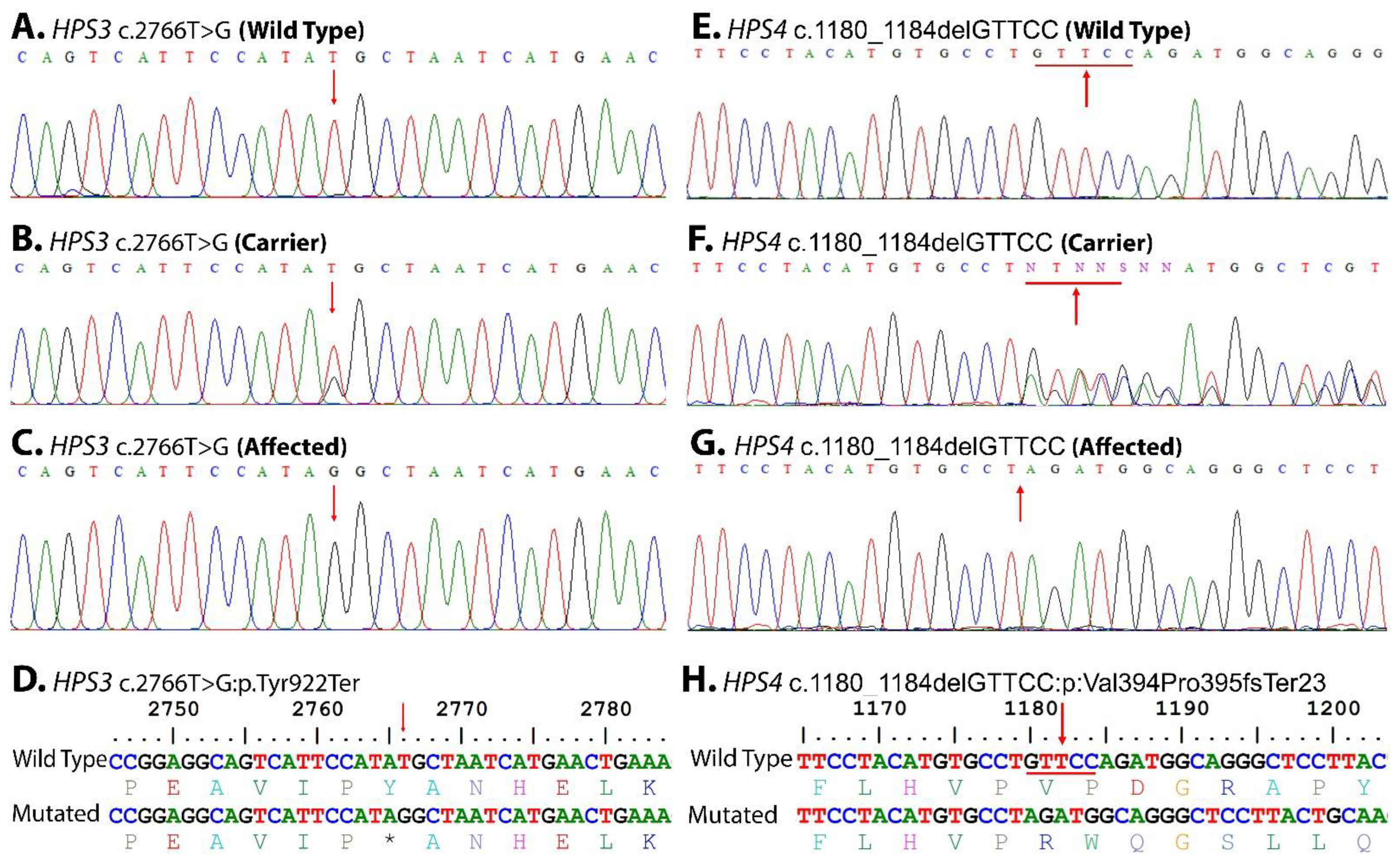
3.2. Genetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huizing, M.; Scher, C.D.; Strovel, E.; Fitzpatrick, D.L.; Hartnell, L.M.; Anikster, Y.; Gahl, W.A. Nonsense mutations in ADTB3A cause complete deficiency of the β3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr. Res. 2002, 51, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Huizing, M.; Malicdan, M.C.; Wang, J.A.; Pri-Chen, H.; Hess, R.A.; Fischer, R.; O’Brien, K.J.; Merideth, M.A.; Gahl, W.A.; Gochuico, B.R. Hermansky–Pudlak syndrome: Mutation update. Hum. Mutat. 2020, 41, 543–580. [Google Scholar] [CrossRef] [PubMed]
- Boeckelmann, D.; Wolter, M.; Käsmann-Kellner, B.; Koehler, U.; Schieber-Nakamura, L.; Zieger, B. A Novel Likely Pathogenic Variant in the BLOC1S5 Gene Associated with Hermansky-Pudlak Syndrome Type 11 and an Overview of Human BLOC-1 Deficiencies. Cells 2021, 10, 2630. [Google Scholar] [CrossRef] [PubMed]
- Toro, J.; Turner, M.; Gahl, W.A. Dermatologic Manifestations of Hermansky-Pudlak Syndrome in Patients With and Without a 16–Base Pair Duplication in the HPS1 Gene. Arch. Dermatol. 1999, 135, 774–780. [Google Scholar] [CrossRef]
- Summers, C.G.; Knobloch, W.H.; Witkop Jr, C.J.; King, R.A. Hermansky-Pudlak syndrome: Ophthalmic findings. Ophthalmology 1988, 95, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Schneier, A.J.; Fulton, A.B. The hermansky-pudlak syndrome: Clinical features and imperatives from an ophthalmic perspective. Semin. Ophthalmol. 2013, 28, 387–391. [Google Scholar] [CrossRef]
- Gahl, W.A.; Brantly, M.; Troendle, J.; Avila, N.A.; Padua, A.; Montalvo, C.; Cardona, H.; Calis, K.A.; Gochuico, B. Effect of pirfenidone on the pulmonary fibrosis of Hermansky–Pudlak syndrome. Mol. Genet. Metab. 2002, 76, 234–242. [Google Scholar] [CrossRef]
- Huizing, M.; Gochuico, B.R.; Gahl, W.A.; Malicdan, M.C.V. Molecular genetics of Hermansky–Pudlak syndrome. eLS 2017, 1–10. [Google Scholar]
- El-Chemaly, S.; O’Brien, K.J.; Nathan, S.D.; Weinhouse, G.L.; Goldberg, H.J.; Connors, J.M.; Cui, Y.; Astor, T.L.; Camp, P.C., Jr.; Rosas, I.O. Clinical management and outcomes of patients with Hermansky-Pudlak syndrome pulmonary fibrosis evaluated for lung transplantation. PLoS ONE 2018, 13, e0194193. [Google Scholar] [CrossRef]
- Hussain, N.; Quezado, M.; Huizing, M.; Geho, D.; White, J.G.; Gahl, W.; Mannon, P. Intestinal disease in Hermansky-Pudlak syndrome: Occurrence of colitis and relation to genotype. Clin. Gastroenterol. Hepatol. 2006, 4, 73–80. [Google Scholar] [CrossRef]
- Felipez, L.; Gokhale, R.; Guandalini, S. Hermansky-Pudlak syndrome: Severe colitis and good response to infliximab. Gastroenterol. Nutr. 2010, 51, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Lowe, G.C.; Guiu, I.S.; Chapman, O.; Rivera, J.; Lordkipanidzé, M.; Dovlatova, N.; Wilde, J.; Watson, S.P.; Morgan, N.V.; UK GAPP Collaborative. Microsatellite markers as a rapid approach for autozygosity mapping in Hermansky-Pudlak syndrome: Identification of the second HPS7 mutation in a patient presenting late in life. Thromb. Haemost. 2013, 109, 766–768. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.A.; Sakuraba, A.; Rubin, D.T. Two complex cases of Hermansky-Pudlak syndrome highlight a potential biologic explanation for an associated Crohn’s disease phenotype. ACG Case Rep. J. 2017, 4, e14. [Google Scholar] [CrossRef]
- Badolato, R.; Prandini, A.; Caracciolo, S.; Colombo, F.; Tabellini, G.; Giacomelli, M.; Cantarini, M.E.; Pession, A.; Bell, C.J.; Dinwiddie, D.L. Exome sequencing reveals a pallidin mutation in a Hermansky-Pudlak–like primary immunodeficiency syndrome. Blood J. Am. Soc. Hematol. 2012, 119, 3185–3187. [Google Scholar] [CrossRef]
- Ammann, S.; Schulz, A.; Krägeloh-Mann, I.; Dieckmann, N.M.; Niethammer, K.; Fuchs, S.; Eckl, K.M.; Plank, R.; Werner, R.; Altmüller, J. Mutations in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood J. Am. Soc. Hematol. 2016, 127, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Fontana, S.; Parolini, S.; Vermi, W.; Booth, S.; Gallo, F.; Donini, M.; Benassi, M.; Gentili, F.; Ferrari, D.; Notarangelo, L.D. Innate immunity defects in Hermansky-Pudlak type 2 syndrome. Blood 2006, 107, 4857–4864. [Google Scholar] [CrossRef]
- Okamura, K.; Abe, Y.; Araki, Y.; Wakamatsu, K.; Seishima, M.; Umetsu, T.; Kato, A.; Kawaguchi, M.; Hayashi, M.; Hozumi, Y. Characterization of melanosomes and melanin in Japanese patients with Hermansky–Pudlak syndrome types 1, 4, 6, and 9. Pigment. Cell Melanoma Res. 2018, 31, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Khan, I.; Khan, M.I.; Latif, M.; Siddiqui, M.I.; Khan, S.U.; Htar, T.T.; Wahid, G.; Ullah, I.; Bibi, F. Whole exome sequencing identifies a novel compound heterozygous GFM1 variant underlying developmental delay, dystonia, polymicrogyria, and severe intellectual disability in a Pakhtun family. Am. J. Med. Genet. Part A 2022, 188, 2693–2700. [Google Scholar] [CrossRef]
- Seo, G.H.; Kim, T.; Choi, I.H.; Park, J.y.; Lee, J.; Kim, S.; Won, D.g.; Oh, A.; Lee, Y.; Choi, J. Diagnostic yield and clinical utility of whole exome sequencing using an automated variant prioritization system, EVIDENCE. Clin. Genet. 2020, 98, 562–570. [Google Scholar] [CrossRef]
- Harrison, C.; Khair, K.; Baxter, B.; Russell-Eggitt, I.; Hann, I.; Liesner, R. Hermansky–Pudlak syndrome: Infrequent bleeding and first report of Turkish and Pakistani kindreds. Arch. Dis. Child. 2002, 86, 297–301. [Google Scholar] [CrossRef]
- Morgan, N.V.; Pasha, S.; Johnson, C.A.; Ainsworth, J.R.; Eady, R.A.; Dawood, B.; McKeown, C.; Trembath, R.C.; Wilde, J.; Watson, S.P. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am. J. Hum. Genet. 2006, 78, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, S.; Shahzad, M.; Tasleem, K.; Sheikh, S.A.; Tariq, N.; Shabbir, A.S.; Ali, M.; Waryah, A.M.; Shaikh, R.S.; Riazuddin, S. Identification and clinical characterization of Hermansky-Pudlak syndrome alleles in the Pakistani population. Pigment. Cell Melanoma Res. 2016, 29, 231. [Google Scholar] [CrossRef] [PubMed]
- Anikster, Y.; Huizing, M.; White, J.; Shevchenko, Y.O.; Fitzpatrick, D.L.; Touchman, J.W.; Compton, J.G.; Bale, S.J.; Swank, R.T.; Gahl, W.A. Mutation of a new gene causes a unique form of Hermansky–Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat. Genet. 2001, 28, 376–380. [Google Scholar] [CrossRef]
- Di Pietro, S.M.; Falcón-Pérez, J.M.; Dell’Angelica, E.C. Characterization of BLOC-2, a complex containing the Hermansky–Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004, 5, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Li, W.; Zhang, Q.; Karim, A.; Novak, E.K.; Sviderskaya, E.V.; Hill, S.P.; Bennett, D.C.; Levin, A.V.; Nieuwenhuis, H.K. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat. Genet. 2002, 30, 321–324. [Google Scholar] [CrossRef]
- Anderson, P.D.; Huizing, M.; Claassen, D.A.; White, J.; Gahl, W.A. Hermansky-Pudlak syndrome type 4 (HPS-4): Clinical and molecular characteristics. Nat. Genet. 2003, 113, 10–17. [Google Scholar] [CrossRef]
- Alfadhel, M.; Umair, M.; Almuzzaini, B.; Alsaif, S.; AlMohaimeed, S.A.; Almashary, M.A.; Alharbi, W.; Alayyar, L.; Alasiri, A.; Ballow, M. Targeted SLC19A3 gene sequencing of 3000 Saudi newborn: A pilot study toward newborn screening. Ann. Clin. Transl. Neurol. 2019, 6, 2097–2103. [Google Scholar] [CrossRef]
- Alyafee, Y.; Al Tuwaijri, A.; Alam, Q.; Umair, M.; Haddad, S.; Alharbi, M.; Ballow, M.; Al Drees, M.; AlAbdulrahman, A.; Al Khaldi, A. Next Generation Sequencing Based Non-invasive Prenatal Testing (NIPT): First Report From Saudi Arabia. Front. Genet. 2021, 12, 630787. [Google Scholar] [CrossRef]
- Alyafee, Y.; Alam, Q.; Tuwaijri, A.A.; Umair, M.; Haddad, S.; Alharbi, M.; Alrabiah, H.; Al-Ghuraibi, M.; Al-Showaier, S.; Alfadhel, M. Next-Generation Sequencing-Based Pre-Implantation Genetic Testing for Aneuploidy (PGT-A): First Report from Saudi Arabia. Genes 2021, 12, 461. [Google Scholar] [CrossRef]
- Alyafee, Y.; Al Tuwaijri, A.; Umair, M.; Alharbi, M.; Haddad, S.; Ballow, M.; Alayyar, L.; Alam, Q.; Althenayyan, S.; Al Ghilan, N. Non-invasive prenatal testing for autosomal recessive disorders: A new promising approach. Front. Genet. 2022, 13, 1047474. [Google Scholar] [CrossRef]



| S/No | Gene | Exon | Primers (5′-3′) | Temp | Product Size (bp) |
|---|---|---|---|---|---|
| 1 | HPS3 | Exon 15 | F 5′-TTGGCAAGTGAGGTTTAGTCTT-3′ | 56 °C | 383 bp |
| R 5′-TTGCAGGAAAGGAAATGAGG-3′ | |||||
| 2 | HPS4 | Exon 11 | F 5′-GCAGGACTGCACAACTCTG-3′ | 57 °C | 637 bp |
| R 5′-CCTTTGACGCAGTGAGTGTA-3′ |
| S/No | Clinicals/Phenotypes with Reported References | HPS3 | HPS4 | ALB-09 | ALB-10 |
|---|---|---|---|---|---|
| 1 | Ocular albinism | + | + | + | + |
| 2 | Reduced visual acuity | + | + | + | + |
| 3 | Horizontal nystagmus | + | + | + | + |
| 4 | Photophobia | + | + | + | + |
| 5 | Iris transillumination | + | + | + | + |
| 6 | Hypopigmentation of retina | + | + | + | + |
| 7 | Foveal hypoplasia | + | + | n/a | n/a |
| 8 | Skin pigment dilution, mild-to-severe, relative to unaffected family members | + | + | + | + |
| 9 | Weakly pigmented basal cell layer | n/a | + | n/a | n/a |
| 10 | Normal number of melanocytes | - | + | n/a | n/a |
| 11 | Reduced amount of melanin pigment in melanocytes | - | + | + | + |
| 12 | Accumulation of ceroid pigment in perivascular macrophages | - | + | + | + |
| 13 | Hair pigment, mild-to-severe, dilution relative to unaffected family members | + | + | + | + |
| 14 | Bleeding tendency | + | + | + | + |
| 15 | Easy bruising | + | + | - | - |
| 16 | Gingival bleeding | + | + | + | + |
| 17 | Epistaxis (nose bleeding) | - | + | + | + |
| 18 | Menorrhagia | + | + | + | + |
| 19 | Absence of platelet-dense bodies | + | + | n/a | n/a |
| 20 | Lack of secondary aggregation response of platelets | + | + | n/a | n/a |
| 21 | Pulmonary fibrosis | - | + | - | + |
| 22 | Restrictive lung disease | - | + | n/a | n/a |
| 23 | Enterocolitis | - | + | + | + |
| 24 | Reported gene | HPS3/BLOC2S1 | HPS4/BLOC3S2 | HPS3/BLOC2S1 | HPS4/BLOC3S2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaman, Q.; Sadeeda; Anas, M.; Rehman, G.; Khan, Q.; Iftikhar, A.; Ahmad, M.; Owais, M.; Ahmad, I.; Muthaffar, O.Y.; et al. Report of Hermansky–Pudlak Syndrome in Two Families with Novel Variants in HPS3 and HPS4 Genes. Genes 2023, 14, 145. https://doi.org/10.3390/genes14010145
Zaman Q, Sadeeda, Anas M, Rehman G, Khan Q, Iftikhar A, Ahmad M, Owais M, Ahmad I, Muthaffar OY, et al. Report of Hermansky–Pudlak Syndrome in Two Families with Novel Variants in HPS3 and HPS4 Genes. Genes. 2023; 14(1):145. https://doi.org/10.3390/genes14010145
Chicago/Turabian StyleZaman, Qaiser, Sadeeda, Muhammad Anas, Gauhar Rehman, Qadeem Khan, Aiman Iftikhar, Mashal Ahmad, Muhammad Owais, Ilyas Ahmad, Osama Yousef Muthaffar, and et al. 2023. "Report of Hermansky–Pudlak Syndrome in Two Families with Novel Variants in HPS3 and HPS4 Genes" Genes 14, no. 1: 145. https://doi.org/10.3390/genes14010145
APA StyleZaman, Q., Sadeeda, Anas, M., Rehman, G., Khan, Q., Iftikhar, A., Ahmad, M., Owais, M., Ahmad, I., Muthaffar, O. Y., Abdulkareem, A. A., Bibi, F., Jelani, M., & Naseer, M. I. (2023). Report of Hermansky–Pudlak Syndrome in Two Families with Novel Variants in HPS3 and HPS4 Genes. Genes, 14(1), 145. https://doi.org/10.3390/genes14010145







