Interacting Networks of the Hypothalamic–Pituitary–Ovarian Axis Regulate Layer Hens Performance
Abstract
1. Introduction
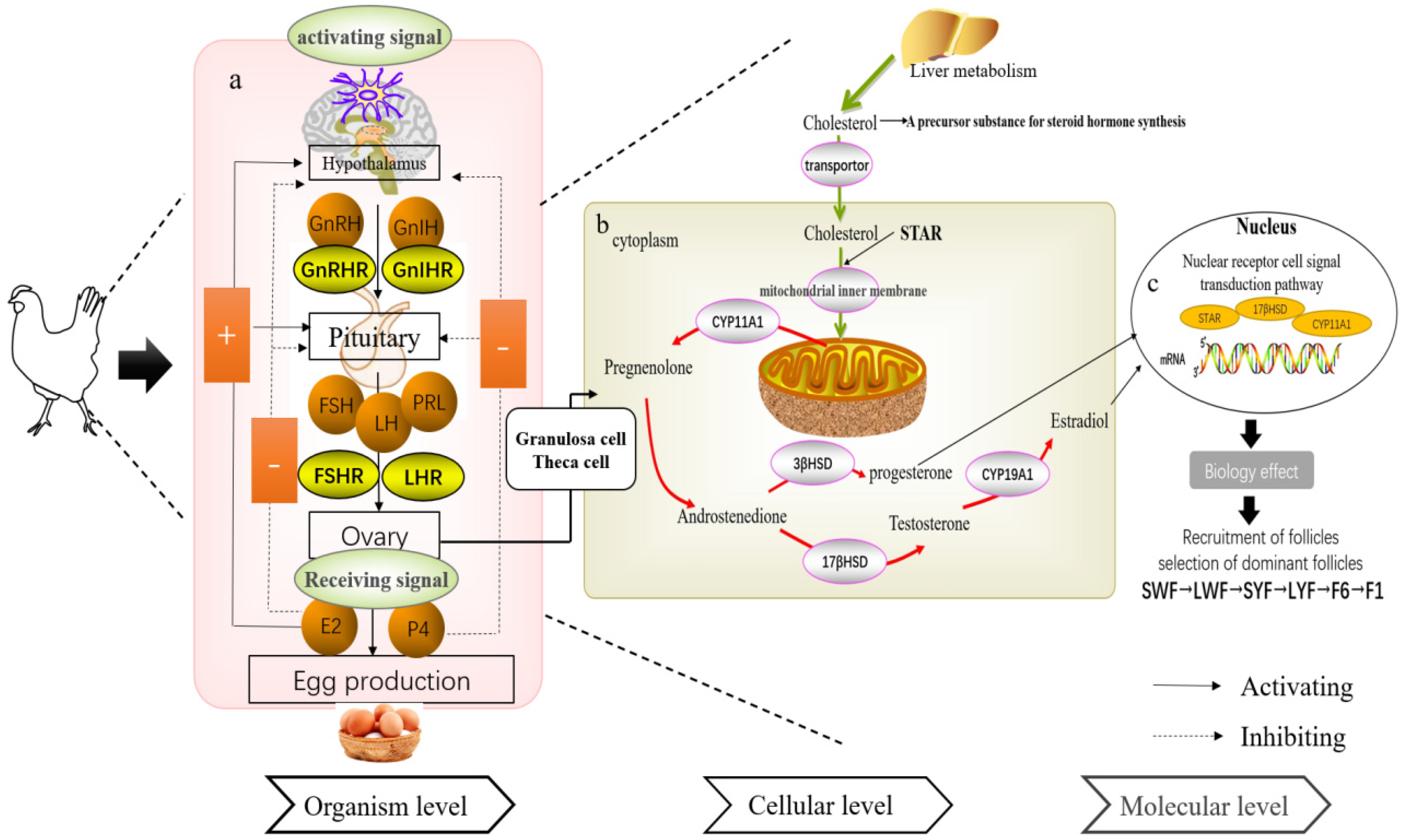
2. Organism-Level Regulation Mechanism of the HPO Axis
2.1. Development Characteristics of the HPO Axis
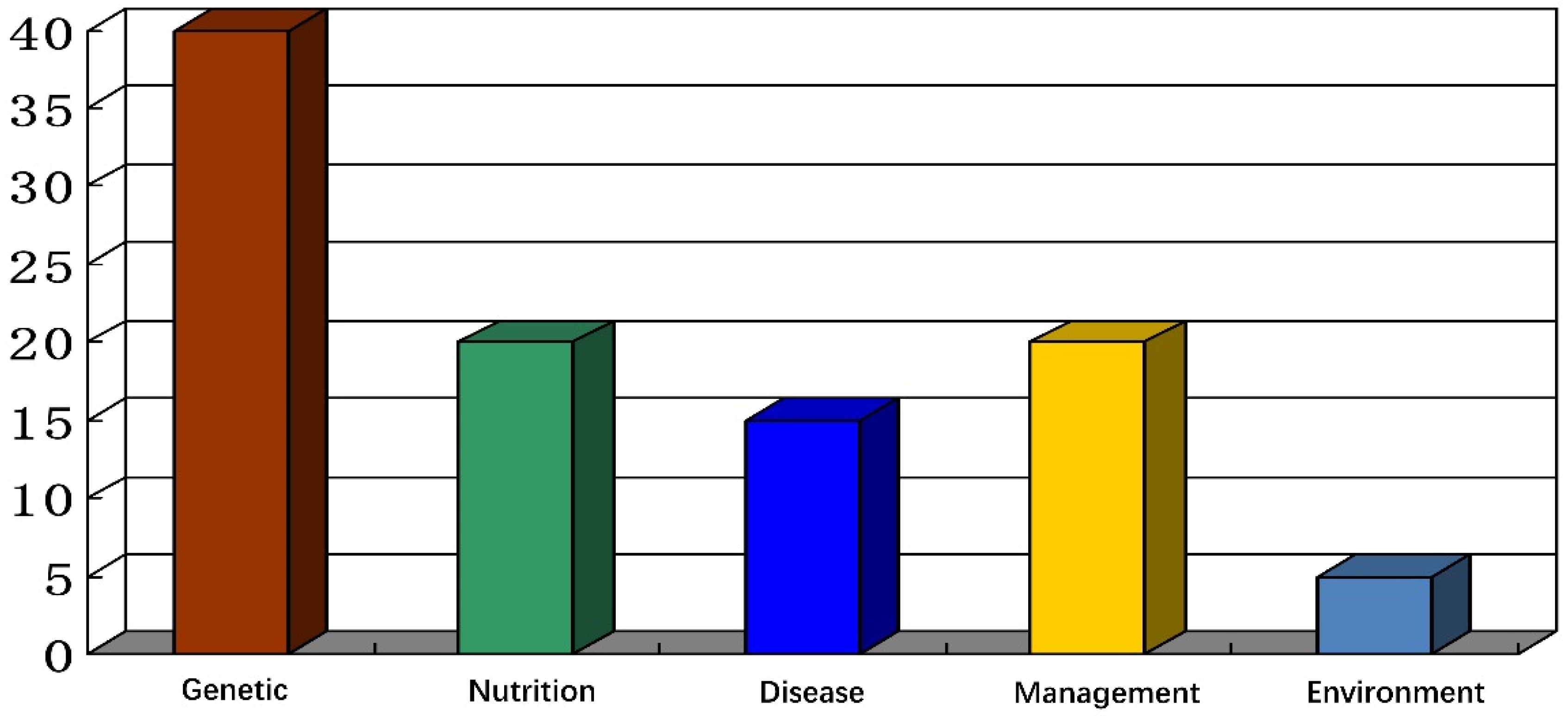
2.2. Factors Affecting the Organism-Level Regulation Mechanism
2.2.1. Factors Related to the Metabolic and Nutritional Status
2.2.2. Environmental Factors
2.2.3. Genetic Factors
3. Cellular-Level Regulation of the HPO Axis in Avians
3.1. Insulin-like Growth Factor
3.2. Epidermal Growth Factor
3.3. Transforming Growth Factor β
3.4. Immune Regulatory Factors
4. Molecular Regulation of the HPO Axis in Avians
4.1. The Role of Reproduction-Related Genes in HPO Axis Development
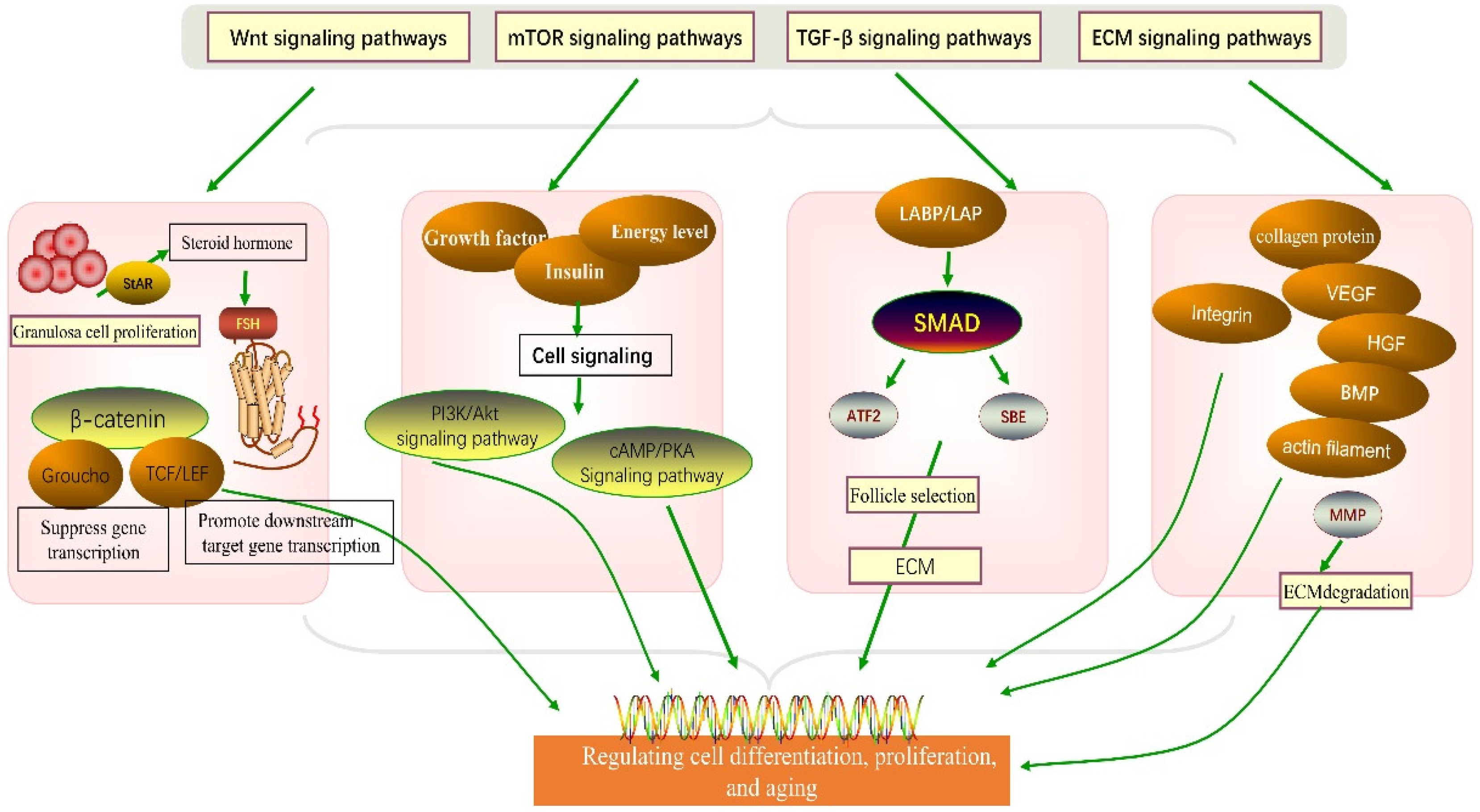
4.2. The Role of Signaling Pathways in Granulosa Cell Development
4.2.1. Wnt Signaling Pathway
4.2.2. mTOR Signaling Pathway
4.2.3. TGF-β Signaling Pathway
4.2.4. ECM Receptor Signaling Pathway
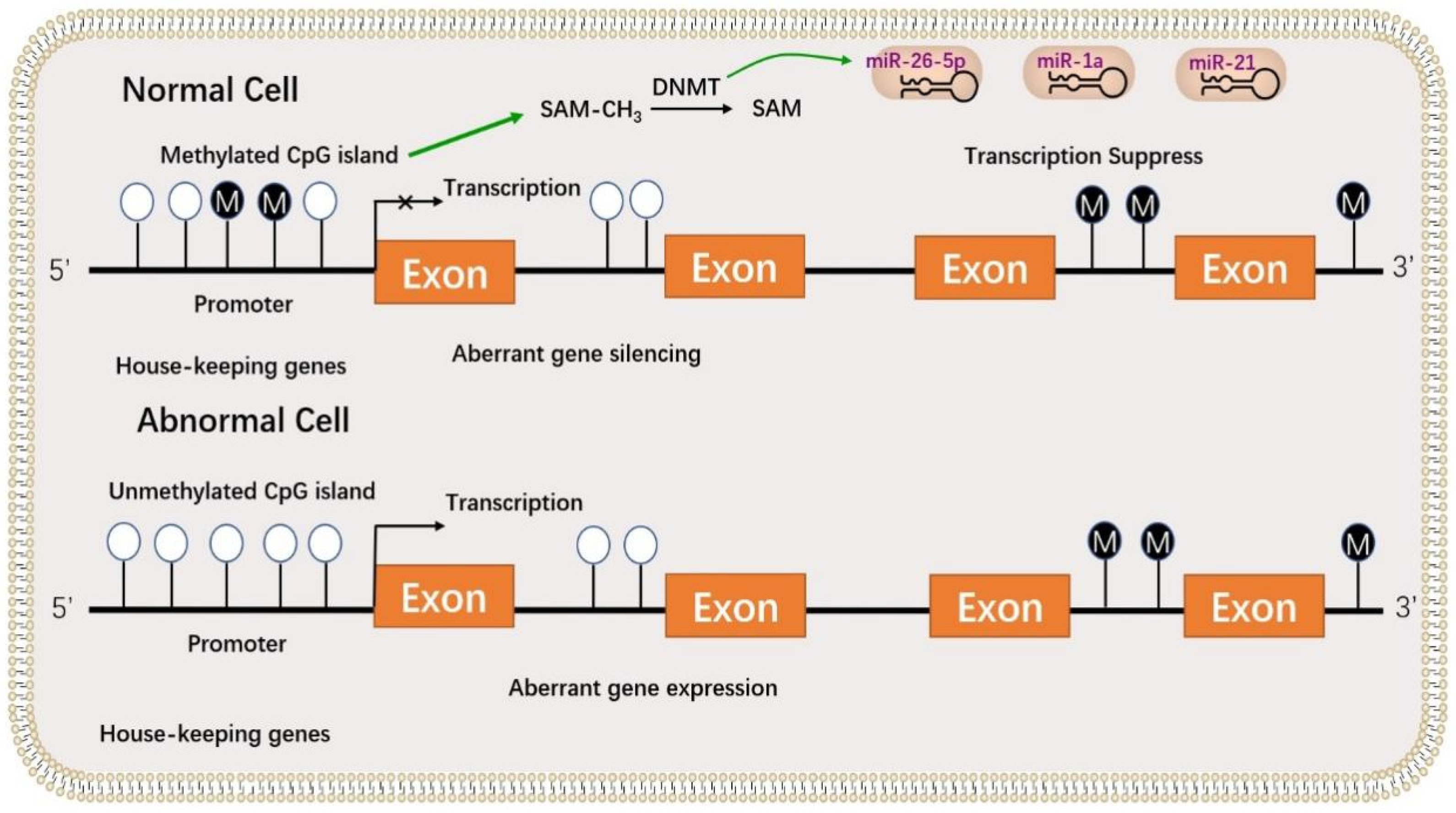
4.3. The Role of Epigenetic Modification in Granulosa Cell Development
4.3.1. The Role of Noncoding RNAs in Granulosa Cell Development
4.3.2. The Role of DNA Methylation in Granulosa Cell Development
5. Interaction Network Regulation Mechanism of the HPO Axis
5.1. Organism-Level Regulation
5.2. Cellular-Level Regulation
5.3. Molecular-Level Regulation
6. Recommendations for Future Research
- (1)
- Compared with the common transcriptome, single-cell RNA sequencing is a high-precision technique that can provide accurate resolution from the multicellular level to a single-cell level. This separates specific characteristics of the reaction cells or a group of cells based on RNA abundance to evaluate the condition of a single cell. Additionally, the single-cell RNA sequencing can be valuable for studying different cell types and developmental track. Therefore, single-cell RNA sequencing is a valuable tool for studying the molecular mechanism of GCs involved in the regulation of the egg production performance, which can effectively improve the efficiency of molecular breeding and accelerate breed selection.
- (2)
- Metabolomics is often referred to as the bridge between genomics and phenotype, and metabolic components can be used as biomarkers for determining complex biological traits. Using the transcriptome data to obtain a large number of differentially expressed genes and differential metabolites obtained through correlation analysis, and from two levels, causes and consequences of organism are analyzed, identifying key gene targets, metabolites, and metabolic pathways to build the key control networks. Efficient seed selection based on biomarkers is also the development direction in the breeding of layer hens.
- (3)
- Proliferation, differentiation, and apoptosis of GCs are the key to prolonging the laying cycle, and GC gene expression is temporally and spatially regulated. Future studies should utilize single-cell RNA sequencing and spatial transcriptomics to analyze the gene expression from both temporal and spatial dimensions in GCs. Moreover, the chromatin accessibility analysis technique can be used to predict the dynamic changes of chromatin conformation during follicle development, which may provide ideas for the breeding of high-reproductive performance hens.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Smabeek, F. Breeding for 500 Eggs in 100 Weeks [EB/OL]. 2 March 2011. Available online: https://www.poultryworld.net/poultry/breeding-for-500-eggs-in-100-weeks/ (accessed on 2 March 2011).
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Announcemengt No.2 of the Ministry of Agricukture of the Peopie’s Republic of China. Available online: http://www.gov.cn/xinwen/202104/29/content_5603727.htm (accessed on 29 April 2021).
- Yang, S.; Chen, J.; Cheng, Y. Characteristics of Livestock Production System in USA and Enlightenment for China. Food Nutr. China 2017, 23, 20–24. [Google Scholar]
- Onagbesan, O.M.; Mast, J.; Goddeeris, B.; Decuypere, E. Effect of TNF-alpha on LH and IGF-I modulated chicken granulosa cell proliferation and progesterone production during follicular development. Reproduction 2000, 120, 433–442. [Google Scholar] [CrossRef][Green Version]
- Lovell, T.M.; Gladwell, R.T.; Groome, N.P.; Knight, P.G. Ovarian follicle development in the laying hen is accompanied by divergent changes in inhibin A, inhibin B, activin A and follistatin production in granulosa and theca layers. J. Endocrinol. 2003, 177, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Rivas, R.E.C.; Nieto, M.P.C.; Kamiyoshi, M. Effects of Steroid Hormone in Avian Follicles. Asian-Australas. J. Anim. Sci. 2016, 29, 487–499. [Google Scholar] [CrossRef]
- Komatsu, K.; Masubuchi, S. Observation of the dynamics of follicular development in the ovary. Reprod. Med. Biol. 2017, 16, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.; Grzesiak, M.; Saito, N.; Kwaśniewska, M.; Sechman, A.; Hrabia, A. Expression of aquaporin 4 in the chicken ovary in relation to follicle development. Reprod. Domest. Anim. 2017, 52, 857–864. [Google Scholar] [CrossRef]
- Brady, K.; Porter, T.E.; Liu, H.C.; Long, J.A. Characterization of the hypothalamo–pituitary–gonadal axis in low and high egg producing turkey hens. Poult. Sci. 2020, 99, 1163–1173. [Google Scholar] [CrossRef]
- Wu, N.; Gaur, U.; Zhu, Q.; Chen, B.; Xu, Z.; Zhao, X.; Yang, M.; Li, D. Expressed microRNA associated with high rate of egg production in chicken ovarian follicles. Anim. Genet. 2017, 48, 205–216. [Google Scholar] [CrossRef]
- Shi, S.; Zhou, X.; Li, J.; Zhang, L.; Hu, Y.; Li, Y.; Yang, G.; Chu, G. MiR-214-3p promotes proliferation and inhibits estradiol synthesis in porcine granulosa cells. J. Anim. Sci. Biotechnol. 2020, 11, 94. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Yao, W.; Du, X.; Li, Q. Lnc2300 is a cis-acting long noncoding RNA of CYP11A1 in ovarian granulosa cells. J. Cell. Physiol. 2022, 237, 4238–4250. [Google Scholar] [CrossRef]
- Kuo, Y.M.; Shiue, Y.L.; Chen, C.F.; Tang, P.C.; Lee, Y.P. Proteomic analysis of hypothalamic proteins of high and low egg production strains of chickens. Theriogenology 2005, 64, 1490–1502. [Google Scholar] [CrossRef]
- Padmanabhan, V.; Karsch, F.J.; Lee, J.S. Hypothalamic, pituitary and gonadal regulation of FSH. Reprod. Suppl. 2002, 59, 67–82. [Google Scholar]
- Cao, C.; Ding, Y.; Kong, X.; Feng, G.; Xiang, W.; Chen, L.; Yang, F.; Zhang, K.; Chu, M.; Wang, P.; et al. Reproductive role of miRNA in the hypothalamic-pituitary axis. Mol. Cell. Neurosci. 2018, 88, 130–137. [Google Scholar] [CrossRef]
- Millar, R.P.; Lu, Z.L.; Pawson, A.J.; Flanagan, C.A.; Morgan, K.; Maudsley, S.R. Gonadotropin-Releasing Hormone Receptors. Endocr. Rev. 2004, 25, 235–275. [Google Scholar] [CrossRef]
- Metallinou, C.; Asimakopoulos, B.; Schröer, A.; Nikolettos, N. Gonadotropin-releasing hormone in the ovary. Reprod. Sci. 2007, 14, 737–749. [Google Scholar] [CrossRef]
- Maddineni, S.; Ocón-Grove, O.M.; Krzysik-Walker, S.M.; Hendricks, G.L., 3rd; Proudman, J.A.; Ramachandran, R. Gonadotrophin-inhibitory hormone receptor expression in the chicken pituitary gland: Potential influence of sexual maturation and ovarian steroids. J. Neuroendocrinol. 2008, 20, 1078–1088. [Google Scholar] [CrossRef]
- Shen, X.; Zeng, H.; Xie, L.; He, J.; Li, J.; Xie, X.; Luo, C.; Xu, H.; Zhou, M.; Nie, Q.; et al. The GTPase Activating Rap/RanGAP Domain-Like 1 Gene Is Associated with Chicken Reproductive Traits. PLoS ONE 2012, 7, e33851. [Google Scholar] [CrossRef]
- Chen, X.; Sun, X.; Chimbaka, I.M.; Qin, N.; Xu, X.; Liswaniso, S.; Xu, R.; Gonzalez, J.M. Transcriptome Analysis of Ovarian Follicles Reveals Potential Pivotal Genes Associated with Increased and Decreased Rates of Chicken Egg Production. Front. Genet. 2021, 12, 622751. [Google Scholar] [CrossRef]
- Robinson, F.E.; Renema, R.A.; Oosterhoff, H.H.; Zuidhof, M.J.; Wilson, J.L. Carcass Traits, Ovarian Morphology and Egg Laying Characteristics in Early Versus Late Maturing Strains of Commercial Egg-Type Hens. Poult. Sci. 2001, 80, 37–46. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Li, Q.; Li, G.; Li, W.; Li, H.; Kang, X.; Tian, Y. Novel Regulatory Factors in the Hypothalamic-Pituitary-Ovarian Axis of Hens at Four Developmental Stages. Front. Genet. 2020, 11, 591672. [Google Scholar] [CrossRef]
- Zhu, G.; Mao, Y.; Zhou, W.; Jiang, Y. Dynamic Changes in the Follicular Transcriptome and Promoter DNA Methylation Pattern of Steroidogenic Genes in Chicken Follicles throughout the Ovulation Cycle. PLoS ONE 2015, 10, e0146028. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.A. Follicle Selection in the Avian Ovary. Reprod. Domest. Anim. 2012, 47, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Woods, D.C. Dynamics of avian ovarian follicle development: Cellular mechanisms of granulosa cell differentiation. Gen. Comp. Endocrinol. 2009, 163, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Qin, N.; Tyasi, T.L.; Zhu, H.; Liu, D.; Yuan, S.; Xu, R. The Hippo/MST Pathway Member SAV1 Plays a Suppressive Role in Development of the Prehierarchical Follicles in Hen Ovary. PLoS ONE 2016, 11, e0160896. [Google Scholar] [CrossRef] [PubMed]
- Robinson, F.E.; Fasenko, G.M.; Renema, R.A. Optimizing Chick Production in Broiler Breeders; Spotted Cow Press: Edmonton, AB, Canada, 2003; Volume 1. [Google Scholar]
- Onagbesan, O.; Bruggeman, V.; Decuypere, E. Intra-ovarian growth factors regulating ovarian function in avian species: A review. Anim. Reprod. Sci. 2009, 111, 121–140. [Google Scholar] [CrossRef]
- Sharp, P.J. Photoperiodic Control of Reproduction in the Domestic Hen1. Poult. Sci. 1993, 72, 897–905. [Google Scholar] [CrossRef]
- Liu, G.; Dunnington, E.A.; Siegel, P.B. Correlated Responses to Long-Term Divergent Selection for Eight-Week Body Weight in Chickens: Growth, Sexual Maturity, and Egg Production. Poult. Sci. 1995, 74, 1259–1268. [Google Scholar] [CrossRef]
- Yang, N.; Jiang, R.S. Recent advances in breeding for quality chickens. Proc. Nutr. Soc. 2005, 61, 373–381. [Google Scholar] [CrossRef]
- Sirotkin, A.V.; Harrath, A.H.; Grossmann, R. The role of metabolic state and obestatin in control of chicken ovarian hormone release. Poult. Sci. 2016, 95, 1939–1942. [Google Scholar] [CrossRef]
- Mehlhorn, J.; Höhne, A.; Baulain, U.; Schrader, L.; Weigend, S.; Petow, S. Estradiol-17ß Is Influenced by Age, Housing System, and Laying Performance in Genetically Divergent Laying Hens (Gallus gallus f.d.). Front. Physiol. 2022, 13, 954399. [Google Scholar] [CrossRef]
- Eusemann, B.K.; Baulain, U.; Schrader, L.; Thöne-Reineke, C.; Patt, A.; Petow, S. Radiographic examination of keel bone damage in living laying hens of different strains kept in two housing systems. PLoS ONE 2018, 13, e0194974. [Google Scholar] [CrossRef]
- Liu, L.; Li, D.; Gilbert, E.R.; Xiao, Q.; Zhao, X.; Wang, Y.; Yin, H.; Zhu, Q. Effect of Monochromatic Light on Expression of Estrogen Receptor (ER) and Progesterone Receptor (PR) in Ovarian Follicles of Chicken. PLoS ONE 2015, 10, e0144102. [Google Scholar] [CrossRef]
- Johnson, A.L.; Solovieva, E.V.; Bridgham, J.T. Relationship between steroidogenic acute regulatory protein expression and progesterone production in hen granulosa cells during follicle development. J. Biol. Reprod. 2002, 67, 1313. [Google Scholar] [CrossRef]
- Zhou, S.; Ma, Y.; Zhao, D.; Mi, Y.; Zhang, C. Transcriptome profiling analysis of underlying regulation of growing follicle development in the chicken. J. Poult. Sci. 2020, 99, 2861–2872. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, L.; Han, K.; Zhang, X.; Zhang, G.; Dai, G.; Wang, J.; Xie, K. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. J. Anim. Reprod. Sci. 2019, 208, 106114. [Google Scholar] [CrossRef]
- Ren, J.; Tian, W.; Jiang, K.; Wang, Z.; Wang, D.; Li, Z.; Yan, F.; Wang, Y.; Tian, Y.; Ou, K.; et al. Global investigation of estrogen-responsive genes regulating lipid metabolism in the liver of laying hens. BMC Genom. 2021, 22, 428. [Google Scholar] [CrossRef]
- Li, H.; Wang, T.; Xu, C.; Wang, D.; Ren, J.; Li, Y.; Tian, Y.; Wang, Y.; Jiao, Y.; Kang, X.; et al. Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genom. 2015, 16, 763. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Cao, J.; Dong, Y.; Wang, Z.; Hu, M.; Chen, Y. Green light inhibits GnRH-I expression by stimulating the melatonin-GnIH pathway in the chick brain. J. Neuroendocr. 2017, 29. [Google Scholar] [CrossRef]
- Zhao, F.Q.; Zhang, Z.W.; Qu, J.P.; Yao, H.D.; Li, M.; Li, S.; Xu, S.W. Cold stress induces antioxidants and Hsps in chicken immune organs. Cell Stress Chaperones 2014, 19, 635–648. [Google Scholar] [CrossRef]
- Farag, M.R.; Alagawany, M. Physiological alterations of poultry to the high environmental temperature. J. Therm. Biol. 2018, 76, 101–106. [Google Scholar] [CrossRef]
- Zhao, X.; Nie, C.; Zhang, J.; Li, X.; Zhu, T.; Guan, Z.; Chen, Y.; Wang, L.; Lv, X.Z.; Yang, W.; et al. Identification of candidate genomic regions for chicken egg number traits based on genome-wide association study. BMC Genom. 2021, 22, 610. [Google Scholar] [CrossRef] [PubMed]
- Ahumada, S.S.M.; Martínez, M.C.G.; Carranza, M.; Ávila, M.J.; Luna, A.J.L.; Harvey, S.; Luna, M.; Arámburo, C. Autocrine/paracrine proliferative effect of ovarian GH and IGF-I in chicken granulosa cell cultures. Gen. Comp. Endocrinol. 2016, 234, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K.; Moteki, H.; Kimura, M.; Ogihara, M. Autocrine secretion of insulin-like growth factor-I mediates growth hormone-stimulated DNA synthesis and proliferation in primary cultures of adult rat hepatocytes. Eur. J. Pharmacol. 2021, 891, 173753. [Google Scholar] [CrossRef] [PubMed]
- Richani, D.; Gilchrist, R.B. The epidermal growth factor network: Role in oocyte growth, maturation and developmental competence. Hum. Reprod. Update 2018, 24, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ye, H.; Hao, L.; Sun, Y.; Li, R.; Li, Y.; Yang, Z. SRSFs mediate the function of AR in the ovarian granulosa cells of patients with PCOS. Genes Dis. 2019, 8, 94–109. [Google Scholar] [CrossRef]
- Chiara Perego, M.; Bellitto, N.; Maylem, E.R.S.; Caloni, F.; Spicer, L.J. Effects of selected hormones and their combination on progesterone and estradiol production and proliferation of feline granulosa cells cultured in vitro. Theriogenology 2021, 168, 1–12. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.; Li, Q.; Liu, J.; Zhang, T.; Zhou, T.; Li, L.; Wang, J.; Xu, H.; He, H. The comprehensive mechanisms underlying nonhierarchical follicular development in geese (Anser cygnoides). Anim. Reprod. Sci. 2015, 159, 131–140. [Google Scholar] [CrossRef]
- Li, H.; Chang, H.M.; Shi, Z.; Leung, P.C.K. The p38 signaling pathway mediates the TGF-β1-induced increase in type I collagen deposition in human granulosa cells. FASEB J. 2020, 34, 15591–15604. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, Q.; Gilbert, E.R.; Cui, Z.; Zhao, X.; Wang, Y.; Yin, H.; Li, D.; Zhang, H.; Zhu, Q. Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 2018, 8, 7231. [Google Scholar] [CrossRef]
- Mishra, S.K.; Chen, B.; Zhu, Q.; Xu, Z.; Ning, C.; Yin, H.; Wang, Y.; Zhao, X.; Fan, X.; Yang, M.; et al. Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci. Rep. 2020, 10, 5976. [Google Scholar] [CrossRef]
- Tamura, K.; Matsushita, M.; Endo, A.; Kutsukake, M.; Kogo, H. Effect of Insulin-Like Growth Factor-Binding Protein 7 on Steroidogenesis in Granulosa Cells Derived from Equine Chorionic Gonadotropin-Primed Immature Rat Ovaries. Biol. Reprod. 2007, 77, 485–491. [Google Scholar] [CrossRef]
- Jiang, R.; Li, J.; Qu, L.; Li, H.; Yang, N. A new single nucleotide polymorphism in the chicken pituitary-specific transcription factor (POU1F1) gene associated with growth rate. Anim. Genet. 2004, 35, 344–346. [Google Scholar] [CrossRef]
- Han, H.; Lei, Q.; Zhou, Y.; Gao, J.; Liu, W.; Li, F.; Zhang, Q.; Lu, Y.; Cao, D. Association between BMP15 Gene Polymorphism and Reproduction Traits and Its Tissues Expression Characteristics in Chicken. PLoS ONE 2015, 10, e0143298. [Google Scholar] [CrossRef]
- Zhou, R.; Miao, Y.; Li, Y.; Li, X.; Xi, J.; Zhang, Z. MicroRNA-150 promote apoptosis of ovine ovarian granulosa cells by targeting STAR gene. Theriogenology 2019, 127, 66–71. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Kiezun, M.; Dobrzyn, K.; Bors, K.; Wyrebek, J.; Kaminska, B.; Kaminski, T.; Smolinska, N. The effect of orexin a on the StAR, CYP11A1 and HSD3B1 gene expression, as well as progesterone and androstenedione secretion in the porcine uterus during early pregnancy and the oestrous cycle. Theriogenology 2020, 143, 179–190. [Google Scholar] [CrossRef]
- Shih, M.C.M.; Chiu, Y.N.; Hu, M.C.; Guo, I.C.; Chung, B. Regulation of steroid production: Analysis of Cyp11a1 promoter. Mol. Cell. Endocrinol. 2011, 336, 80–84. [Google Scholar] [CrossRef]
- Chien, Y.; Cheng, W.C.; Wu, M.R.; Jiang, S.T.; Shen, C.K.J.; Chung, B. Misregulated Progesterone Secretion and Impaired Pregnancy in Cyp11a1 Transgenic Mice1. Biol. Reprod. 2013, 89, 91. [Google Scholar] [CrossRef]
- Chen, P.; Cescon, M.; Bonaldo, P. Collagen VI in cancer and its biological mechanisms. Trends Mol. Med. 2013, 19, 410–417. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Akbari, D.N.; Shoorei, H.; Sharifi, G.; Mohaqiq, M.; Majidpoor, J.; Dinger, M.E.; Taheri, M.; Ghafouri, F.S. Non-coding RNAs modulate function of extracellular matrix proteins. Biomed. Pharmacother. 2021, 136, 111240. [Google Scholar] [CrossRef] [PubMed]
- Umer, S.; Sammad, A.; Zou, H.; Khan, A.; Weldegebriall, S.B.; Hao, H.; Zhao, X.; Wang, Y.; Zhao, S.; Zhu, H. Regulation of AMH, AMHR-II, and BMPs (2,6) Genes of Bovine Granulosa Cells Treated with Exogenous FSH and Their Association with Protein Hormones. Genes 2019, 10, 1038. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Angelini, C.; De Feis, I.; Ciccodicola, A. Uncovering the Complexity of Transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010, 2010, 853916. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fu, L.; Xue, F.; Li, Y.; Xu, H.; Chen, J. Digital gene expression profiling and validation study highlight Cyclin F as an important regulator for sperm motility of chickens. Poult. Sci. 2019, 98, 5118–5126. [Google Scholar] [CrossRef]
- Matulova, M.; Rajova, J.; Vlasatikova, L.; Volf, J.; Stepanova, H.; Havlickova, H.; Sisak, F.; Rychlik, I. Characterization of Chicken Spleen Transcriptome after Infection with Salmonella enterica Serovar Enteritidis. PLoS ONE 2012, 7, e48101. [Google Scholar] [CrossRef]
- Li, Q.; Wang, N.; Du, Z.; Hu, X.; Chen, L.; Fei, J.; Wang, Y.; Li, N. Gastrocnemius transcriptome analysis reveals domestication induced gene expression changes between wild and domestic chickens. Genomics 2012, 100, 314–319. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Liu, Z.; Guo, X.; Du, Y.; Yuan, Z.; Guo, M.; Kang, L.; Sun, Y.; Jiang, Y. Transcriptome Analysis on Single Small Yellow Follicles Reveals That Wnt4 Is Involved in Chicken Follicle Selection. Front. Endocrinol. 2017, 8, 317. [Google Scholar] [CrossRef]
- Shen, M.; Li, T.; Zhang, G.; Wu, P.; Chen, F.; Lou, Q.; Chen, L.; Yin, X.; Zhang, T.; Wang, J. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genom. 2019, 20, 96. [Google Scholar] [CrossRef]
- Magaway, C.; Kim, E.; Jacinto, E. Targeting mTOR and Metabolism in Cancer: Lessons and Innovations. Cells 2019, 8, 1584. [Google Scholar] [CrossRef]
- Liu, J.; Wu, D.C.; Qu, L.H.; Liao, H.Q.; Li, M.X. The role of mTOR in ovarian Neoplasms, polycystic ovary syndrome, and ovarian aging. Clin. Anat. 2018, 31, 891–898. [Google Scholar] [CrossRef]
- Moschetta, M.; Reale, A.; Marasco, C.; Vacca, A.; Carratù, M.R. Therapeutic targeting of the mTOR-signalling pathway in cancer: Benefits and limitations. Br. J. Pharmacol. 2014, 171, 3801–3813. [Google Scholar] [CrossRef]
- Ardestani, A.; Lupse, B.; Kido, Y.; Leibowitz, G.; Maedler, K. mTORC1 Signaling: A Double-Edged Sword in Diabetic β Cells. Cell Metab. 2018, 27, 314–331. [Google Scholar] [CrossRef]
- Perluigi, M.; Di Domenico, F.; Butterfield, D.A. mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis. 2015, 84, 39–49. [Google Scholar] [CrossRef]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Ocón-Grove, O.M.; Poole, D.H.; Johnson, A.L. Bone morphogenetic protein 6 promotes FSH receptor and anti-Müllerian hormone mRNA expression in granulosa cells from hen prehierarchal follicles. Reproduction 2012, 143, 825–833. [Google Scholar] [CrossRef][Green Version]
- Hao, E.Y.; Wang, D.H.; Chen, Y.F.; Zhou, R.Y.; Chen, H.; Huang, R.L. The relationship between the mTOR signaling pathway and ovarian aging in peak-phase and late-phase laying hens. Poult. Sci. 2021, 100, 334–347. [Google Scholar] [CrossRef]
- Fabregat, I.; Moreno, C.J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Dijke, P. TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef]
- Batlle, E.; Massagué, J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Roberts, A.B.; Derynck, R. Meeting Report: Signaling Schemes for TGF-β. Sci. STKE 2001, 2001, pe43. [Google Scholar] [CrossRef]
- Moustakas, A.; Pardali, K.; Gaal, A.; Heldin, C.H. Mechanisms of TGF-β signaling in regulation of cell growth and differentiation. Immunol. Lett. 2002, 82, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ma, Y.; Yao, J.; Zhao, A.; Xie, C.; Mi, Y.; Zhang, C. TGF-β1-induced collagen promotes chicken ovarian follicle development via an intercellular cooperative pattern. Cell Biol. Int. 2021, 45, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Irving, R.H.F.; Rodgers, R.J. Extracellular matrix in ovarian follicular development and disease. Cell Tissue Res. 2005, 322, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O.; Naba, A. Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.Y.; Lim, J.O.; Lee, E.H.; Han, M.H.; Ha, Y.S.; Lee, J.N.; Kim, B.S.; Park, M.J.; Yeo, M.; Jung, B.; et al. Preparation and Characterization of Human Adipose Tissue-Derived Extracellular Matrix, Growth Factors, and Stem Cells: A Concise Review. Tissue Eng. Regen. Med. 2019, 16, 385–393. [Google Scholar] [CrossRef]
- Hrabia, A.; Wolak, D.; Sechman, A. Response of the matrix metalloproteinase system of the chicken ovary to prolactin treatment. Theriogenology 2021, 169, 21–28. [Google Scholar] [CrossRef]
- Jathar, S.; Kumar, V.; Srivastava, J.; Tripathi, V. Technological Developments in lncRNA Biology. Adv. Exp. Med. Biol. 2017, 1008, 283–323. [Google Scholar] [CrossRef]
- Gebremedhn, S.; Salilew-Wondim, D.; Ahmad, I.; Sahadevan, S.; Hossain, M.M.; Hoelker, M.; Rings, F.; Neuhoff, C.; Tholen, E.; Looft, C.; et al. MicroRNA Expression Profile in Bovine Granulosa Cells of Preovulatory Dominant and Subordinate Follicles during the Late Follicular Phase of the Estrous Cycle. PLoS One 2015, 10, e0125912. [Google Scholar] [CrossRef]
- Zielak-Steciwko, A.E.; Browne, J.A.; McGettigan, P.A.; Gajewska, M.; Dzięcioł, M.; Szulc, T.; Evans, A.C. Expression of microRNAs and their target genes and pathways associated with ovarian follicle development in cattle. Physiol. Genom. 2014, 46, 735–745. [Google Scholar] [CrossRef]
- Yin, H.; Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 2007, 450, 304–308. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, L.; Li, Y.; Jiang, P.; Wang, Y.; Wang, P.; Kang, L.; Wang, Y.; Sun, Y.; Jiang, Y. Identification and characterization of microRNA in the lung tissue of pigs with different susceptibilities to PCV2 infection. Vet. Res. 2018, 49, 18. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Ding, N.; Teng, J.; Zhang, S.; Zhang, Q.; Tang, H. miR-34a regulates adipogenesis in porcine intramuscular adipocytes by targeting ACSL4. BMC Genet. 2020, 21, 33. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.K.; Yatskievych, T.A.; Antin, P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef]
- Hicks, J.A.; Trakooljul, N.; Liu, H.C. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol. Genom. 2010, 41, 185–193. [Google Scholar] [CrossRef]
- Wu, H.; Fan, F.; Liang, C.; Zhou, Y.; Qiao, X.; Sun, Y.; Jiang, Y.; Kang, L. Variants of pri-miR-26a-5p polymorphisms are associated with values for chicken egg production variables and affects abundance of mature miRNA. Anim. Reprod. Sci. 2019, 201, 93–101. [Google Scholar] [CrossRef]
- Kang, L.; Yang, C.; Wu, H.; Chen, Q.; Huang, L.; Li, X.; Tang, H.; Jiang, Y. miR-26a-5p Regulates TNRC6A Expression and Facilitates Theca Cell Proliferation in Chicken Ovarian Follicles. DNA Cell Biol. 2017, 36, 922–929. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Zhu, Q.; Xie, X.; Yang, H.; Wang, J.; Pian, H.; Yu, D. Study on the Effect of WNT6 Gene on Granulosa Cells of Laying Hens and Its Mechanism. J. Nanjing Agric. Univ. 2021, 44. [Google Scholar]
- He, H.; Li, D.; Tian, Y.; Wei, Q.; Amevor, F.K.; Sun, C.; Yu, C.; Yang, C.; Du, H.; Jiang, X.; et al. miRNA sequencing analysis of healthy and atretic follicles of chickens revealed that miR-30a-5p inhibits granulosa cell death via targeting Beclin1. J. Anim. Sci. Biotechnol. 2022, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Xue, H.; Sun, C.; Li, J.; He, H.; Amevor, F.K.; Tan, B.; Ma, M.; Tian, K.; Zhang, Z.; et al. Gga-miR-146b-3p promotes apoptosis and attenuate autophagy by targeting AKT1 in chicken granulosa cells. Theriogenology 2022, 190, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Teng, J.; Han, X.; Zhang, S.; Zhang, Q.; Tang, H. miR-458b-5p regulates ovarian granulosa cells proliferation through Wnt/β-catenin signaling pathway by targeting catenin beta-1. Anim. Biosci. 2021, 34, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Li, J.; He, H.; Cao, Y.; Li, D.; Amevor, F.K.; Zhang, Y.; Wang, J.; Yu, C.; Yang, C.; et al. miR-23b-3p inhibits chicken granulosa cell proliferation and steroid hormone synthesis via targeting GDF9. Theriogenology 2022, 177, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Ocłoń E, Hrabia A. miRNA expression profile in chicken ovarian follicles throughout development and miRNA-mediated MMP expression. Theriogenology 2021, 160, 116–127. [CrossRef]
- Wu, X.; Zhang, N.; Li, J.; Zhang, Z.; Guo, Y.; Li, D.; Zhang, Y.; Gong, Y.; Jiang, R.; Li, H.; et al. gga-miR-449b-5p Regulates Steroid Hormone Synthesis in Laying Hen Ovarian Granulosa Cells by Targeting the IGF2BP3 Gene. Animals 2022, 12, 2710. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, J.; Lei, Q.; Han, H.; Liu, W.; Cunwei, T.; Li, F.; Cao, D. Transcriptome Analysis of the Chicken Follicular Theca Cells with miR-135a-5p Suppressed. G3 2020, 10, 4071–4081. [Google Scholar] [CrossRef]
- Yu, C.; Qiu, M.; Yin, H.; Zhang, Z.; Hu, C.; Jiang, X.; Du, H.; Li, Q.; Li, J.; Xiong, X.; et al. miR-138-5p promotes chicken granulosa cell apoptosis viatargeting SIRT1. Anim. Biotechnol. 2022, 6, 1–10. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; He, H.; Zhang, Y.; He, H.; Sun, C.; Zhang, X.; Wang, X.; Kan, Z.; Su, Y.; et al. miR-10a-5p inhibits chicken granulosa cells proliferation and Progesterone(P4) synthesis by targeting MAPRE1 to suppress CDK2. Theriogenology 2022, 192, 97–108. [Google Scholar] [CrossRef]
- Zhang, G.; Cui, Z.; Li, J.; Zhang, D.; Li, Z.; Lin, Z.; Yin, H.; Ran, J.; Wang, Y.; Liu, Y. miR-122-5p regulates proliferation and apoptosis of chicken granulosa cells of hierarchal follicles by targeting MAPK3. Gene 2022, 824, 146397. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.; Zhang, H.; Han, H.; Yang, J.; Li, W.; Wang, K. Transcriptome Analysis Reveals miR-302a-3p Affects Granulosa Cell Proliferation by Targeting DRD1 in Chickens. Front. Genet. 2022, 13, 832762. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez, P.V.; Qu, K.; Zheng, G.X.; Chow, J.; Kim, G.E. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 2013, 108, 2419–2425. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, H.; Zhang, G.; Wu, P.; Chen, L.; Shen, M.; Li, T.; Lv, X.; Gu, Y.; Wang, J. Transcriptome analysis of long noncoding RNAs and mRNAs in granulosa cells of Jinghai Yellow chickens illuminated with red light. Front. Genet. 2021, 12, 104. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Zi, C.; Wang, Z. Transcriptomic analysis of the red and green light responses in Columba livia domestica. 3 Biotech 2019, 9, 20. [Google Scholar] [CrossRef]
- Peng, Y.; Chang, L.; Wang, Y.; Wang, R.; Hu, L.; Zhao, Z.; Geng, L.; Liu, Z.; Gong, Y.; Li, J.; et al. Genome-wide differential expression of long noncoding RNAs and mRNAs in ovarian follicles of two different chicken breeds. Genomics 2019, 111, 1395–1403. [Google Scholar] [CrossRef]
- Xu, X.F.; Li, J.; Cao, Y.X.; Chen, D.W.; Zhang, Z.G.; He, X.J.; Ji, D.M.; Chen, B.L. Differential Expression of Long Noncoding RNAs in Human Cumulus Cells Related to Embryo Developmental Potential: A Microarray Analysis. Reprod. Sci. 2015, 22, 672–678. [Google Scholar] [CrossRef]
- Zhong, C.; Liu, Z.; Qiao, X.; Kang, L.; Sun, Y.; Jiang, Y. Integrated transcriptomic analysis on small yellow follicles reveals that sosondowah ankyrin repeat domain family member A inhibits chicken follicle selection. Anim. Biosci. 2021, 34, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xia, T.; Li, H.; Li, Z.; Sun, G.; Li, G.; Tian, Y.; Liu, X.; Xu, D.; Kang, X. Estrogen enhances the expression of a growth-associated long noncoding RNA in chicken liver via ERα. Br. Poult. Sci. 2021, 62, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, J.; Sun, Y.; Li, Y.; Wang, P.; Shi, L.; Ni, A.; Zong, Y.; Zhao, J.; Bian, S.; et al. Genetic Basis of Sexual Maturation Heterosis: Insights from Ovary lncRNA and mRNA Repertoire in Chicken. Front. Endocrinol. 2022, 13, 951534. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, W.; Wang, X.; Dang, Y.; Xu, L.; Lu, G.; Chan, W.Y.; Leung, P.C.K.; Zhao, S.; Qin, Y. lncRNA DDGC participates in premature ovarian insufficiency through regulating RAD51 and WT1. Mol. Ther. Nucleic Acids 2021, 26, 1092–1106. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, H.; Pi, J.; Zhang, H.; Pan, A.; Pu, Y.; Liang, Z.; Shen, J.; Du, J. The circular RNA aplacirc_13267 upregulates duck granulosa cell apoptosis by the apla-miR-1-13/THBS1 signaling pathway. J. Cell. Physiol. 2020, 235, 5750–5763. [Google Scholar] [CrossRef]
- Lu, X.; Gao, H.; Zhu, B.; Lin, G. Circular RNA circ_RANBP9 exacerbates polycystic ovary syndrome via microRNA-136-5p/XIAP axis. Bioengineered 2021, 12, 6748–6758. [Google Scholar] [CrossRef]
- Li, J. circ EML1 Modulates Steroid Synthesis and Secretion of Hen’s Follicular Granulosa Cells via Targeting the gga-mi R-449a/IGF2BP3 axis. Ph.D. Thesis, Henan Agricultural University, Henan, China, 2022. [Google Scholar]
- Wang, Y.; Guo, Z.; Zi, C.; Wu, P.; Lv, X.; Chen, L.; Chen, F.; Zhang, G.; Wang, J. CircRNA expression in chicken granulosa cells illuminated with red light. Poult. Sci. 2022, 101, 101734. [Google Scholar] [CrossRef]
- Ma, M.; Wang, H.; Zhang, Y.; Zhang, J.; Liu, J.; Pan, Z. circRNA-Mediated Inhibin-Activin Balance Regulation in Ovarian Granulosa Cell Apoptosis and Follicular Atresia. Int. J. Mol. Sci. 2021, 22, 9113. [Google Scholar] [CrossRef]
- Shen, M.; Li, T.; Chen, F.; Wu, P.; Wang, Y.; Chen, L.; Xie, K.; Wang, J.; Zhang, G. Transcriptomic Analysis of circRNAs and mRNAs Reveals a Complex Regulatory Network That Participate in Follicular Development in Chickens. Front. Genet. 2020, 11, 503. [Google Scholar] [CrossRef]
- Mehta, A.; Dobersch, S.; Romero, O.A.J.; Barreto, G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015, 34, 229–241. [Google Scholar] [CrossRef]
- Elhamamsy, A.R. DNA methylation dynamics in plants and mammals: Overview of regulation and dysregulation. Cell Biochem. Funct. 2016, 34, 289–298. [Google Scholar] [CrossRef]
- Law, P.P.; Holland, M.L. DNA methylation at the crossroads of gene and environment interactions. Essays Biochem. 2019, 63, 717–726. [Google Scholar] [CrossRef]
- Chapman, A.G.; Cotton, A.M.; Kelsey, A.D.; Brown, C.J. Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 2014, 15, 89. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA Methylation, Its Mediators and Genome Integrity. Int. J. Biol. Sci. 2015, 11, 604–617. [Google Scholar] [CrossRef]
- Chambers, S.; Tarasov, K.; Aon, M.A.; Lammons, J.; Chakir, K.; Tarasova, Y.; Lukyanenko, Y.; Lyashkov, A.; Lakatta, E. Metabolic Enzyme Acetylation is Elicited in Response to Stress Induced by Cardiac Specific Overexpression of Human ADCY8. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Xin, Y.; Ning, S.; Zhang, L.; Cui, M. CDC27 Facilitates Gastric Cancer Cell Proliferation, Invasion and Metastasis via Twist-Induced Epithelial-Mesenchymal Transition. Cell. Physiol. Biochem. 2018, 50, 501–511. [Google Scholar] [CrossRef]
- Abbaspourkharyeki, M.; Anvekar, N.J.; Ramachandra, N.B. The Possible Role of Point Mutations and Activation of the CDC27 Gene in Progression of Multiple Myeloma. Meta Gene 2020, 26, 100761. [Google Scholar] [CrossRef]
- Qian, U.; Li, G.; Yin, J.; Zhang, H.; Zhou, C.; Zhu, Y.; Xing, W.J. Screening of Genes with Differential Methylated CpG Island Related to Inbreeding Depression of Chicken Reproduction. Acta Vet. Zootech. Sin. 2021, 52, 943–953. [Google Scholar]
- Barua, A.; Bitterman, P.; Abramowicz, J.S.; Dirks, A.L.; Bahr, J.M.; Hales, D.B.; Bradaric, M.J.; Edassery, S.L.; Rotmensch, J.; Luborsky, J.L. Histopathology of Ovarian Tumors in Laying Hens: A Preclinical Model of Human Ovarian Cancer. Int. J. Gynecol. Cancer 2009, 19, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeong, W.; Lim, W.; Lim, C.H.; Bae, S.M.; Kim, J.; Bazer, F.W.; Song, G. Hypermethylation and Post-Transcriptional Regulation of DNA Methyltransferases in the Ovarian Carcinomas of the Laying Hen. PLoS ONE 2013, 8, e61658. [Google Scholar] [CrossRef]
- Dvorská, D.; Braný, D.; Nagy, B.; Grendár, M.; Poka, R.; Soltész, B.; Jagelková, M.; Zelinová, K.; Lasabová, Z.; Zubor, P.; et al. Aberrant Methylation Status of Tumour Suppressor Genes in Ovarian Cancer Tissue and Paired Plasma Samples. Int. J. Mol. Sci. 2019, 20, 4119. [Google Scholar] [CrossRef]
- Sendžikaitė, G.; Hanna, C.W.; Stewart, M.K.R.; Ivanova, E.; Kelsey, G. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019, 10, 1884. [Google Scholar] [CrossRef]
| miRNA | Target Genes | Signaling Pathways | Function | Tissue | Reference |
|---|---|---|---|---|---|
| miR-29c-3p | FOXL2 | cAMP/focal adhesion | Transcription factor Promotes differentiation of pre-hierarchical GCs (PhGCs) and preovulatory GCs (PoGCs) | Chicken (PhGCs/PoGCs) | [71] |
| / | WNT6/LRP1/FOXO1 | Wnt/β-catenin dependent pathway | WNT6 cooperates with FSH to promote follicle development | Hyline-brown (SYF) | [103] |
| gga-miR-30a-5p | BCL1 | miR-30a-5p regulatory pathway | Activates (proliferation) | Chicken | [104] |
| gga-miR-146b-3p | AKT1 | PI3K/ATK | Inhibits (proliferation) | Chicken | [105] |
| miR-458b-5p | CTNNB1 (3′UTR) | Wnt/β-catenin | Inhibits (proliferation) | Chicken (Hyline Brown ) | [106] |
| miR-26a-5p | TNRC6A | / | facilitates chicken ovarian cell proliferation | Chicken (ovarian) | [100] |
| miR-23b-3p | GDF9 | TGF-β signaling pathway | Inhibits (proliferation) steroid hormone synthesis | Chicken (SYF) | [107] |
| miR-200a | ZEB1/SIP1/SIRT1 | reproduction regulation pathways | Inhibit granulosa cells proliferation | Lu hua chicken | [10] |
| miR-138-1-3p | COL4A5 | matrix metalloproteinase (MMP) | Remodeling of the extracellular matrix during ovarian follicle development in chickens | Chicken (largest preovulatory follicles) | [108] |
| miR-449b-5p | IGF2BP3 | Steroid hormone synthesis signaling pathway | Regulate the expression of key steroidogenesis-related genes (StAR and CYP19A1) | Chicken (SYF) | [109] |
| miR-135a-5p | KLF4/ATP8A1/CPLX1 | p53 Signaling pathway | Involve in proliferation and differentiation in chicken ovarian follicular | Chicken | [110] |
| miR-138-5p | SIRT1 | Apoptosis signaling pathway | Promotes apoptosis and follicular atresia | Chicken | [111] |
| miR-10a-5p | MAPRE1 | / | Inhibits proliferation and progesterone synthesis | Tian fu chicken | [112] |
| miR-122-5p | MAPK3 | / | Promotes apoptosis through the post-transcriptional downregulation of MAPK3. | Chicken (hierarchal follicles) | [113] |
| miR-302a-3p | DRD1 | / | Inhibits GCs proliferation | Chicken (Small yellow follicle) | [114] |
| LncRNA | Target Gene | Function | Species | Reference |
|---|---|---|---|---|
| LncRNAXLOC_110025 | MOS BMP15 WNT6 CDC25A | Promote follicle development | Hy-line Brown | [122] |
| LncRNA_138134 | PSMD6 | Affect oocyte maturation | Human (GCs) | [123] |
| LncRNA_210520.2 | SOWAHA | Inhibited the proliferation of granulosa cells | Chicken (SYF) | [124] |
| Lnc RNAGLM | ERα | Regulated by estrogen through ERα | Laying hens | [125] |
| LncRNALTR | ERβ | Involved in lipid metabolism | Chicken | [10] |
| LncRAN MTSRG.17017.1 MTSRG.6475.20 | CACNA1C TGFB1 | Affect gonad development and GnRH signaling pathway | White Leghorns and Beijing You chickens | [126] |
| LncRNA XLOC_001347 XLOC_016063 XLOC_02660 XLOC_03201 XLOC_005141 | CASP6/MMP2/SMAD2 | Involved cell growth, proliferation, and development | Broody chickens (BC) and normal ovaries (NO) | [52] |
| LncRNA RP4-545C24.1 | RAD51 WT1 | Inhibition of DNA damage repair capacity | Human | [127] |
| CircRNA | miRNA | Target Gene | Function | Species | Reference |
|---|---|---|---|---|---|
| circRNA-aplacirc_13267 | apla-miR-1-13 | THBS1 | Promotes granulosa cell apoptosis | Duck | [131] |
| circRNA_RANBP9 | miR-136-5p | XIAP | Inhibits granulosa cell apoptosis | Human (Polycystic ovary syndrome) | [132] |
| circRNA_EML1 | miR-449a | IGF2BP3 | Promotes granulosa cell steroid hormone synthesis and estrogen and progesterone secretion, down regulate miR-449a | Hyline Brown | [133] |
| circRNA_0320 circRNA_0185 | miR-143-3p | FSHR | Promotes GC differentiation and follicle development | Chicken (SYF) | [134] |
| sss-circINHA-001 | miR-24-5p miR-7144-3p miR-9830-5p | INHA | Resisting GC apoptosis and follicular atresia | Chicken | [135] |
| circRNA_8:6369673|6402248 circRNA_8:6369673|642209 circRNA_8:6384248|6402248 | miR-1625-3p miR-1552-3p miR-16-2-3p miR-18b-3p miR-200a-3p | RalGPS2 | Regulate GC development | Chicken (SYF/F6/F1) | [72] |
| novel_circ0004730 | / | ESR | Cell growth, proliferation, differentiation, and apoptosis | Chicken (SYF/F6/F1) | [136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Pan, H.; Liu, Y.; He, Y.; Shi, H.; Ge, C. Interacting Networks of the Hypothalamic–Pituitary–Ovarian Axis Regulate Layer Hens Performance. Genes 2023, 14, 141. https://doi.org/10.3390/genes14010141
Zhao J, Pan H, Liu Y, He Y, Shi H, Ge C. Interacting Networks of the Hypothalamic–Pituitary–Ovarian Axis Regulate Layer Hens Performance. Genes. 2023; 14(1):141. https://doi.org/10.3390/genes14010141
Chicago/Turabian StyleZhao, Jinbo, Hongbin Pan, Yong Liu, Yang He, Hongmei Shi, and Changrong Ge. 2023. "Interacting Networks of the Hypothalamic–Pituitary–Ovarian Axis Regulate Layer Hens Performance" Genes 14, no. 1: 141. https://doi.org/10.3390/genes14010141
APA StyleZhao, J., Pan, H., Liu, Y., He, Y., Shi, H., & Ge, C. (2023). Interacting Networks of the Hypothalamic–Pituitary–Ovarian Axis Regulate Layer Hens Performance. Genes, 14(1), 141. https://doi.org/10.3390/genes14010141





