Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India
Abstract
1. Introduction
2. Materials and Methods
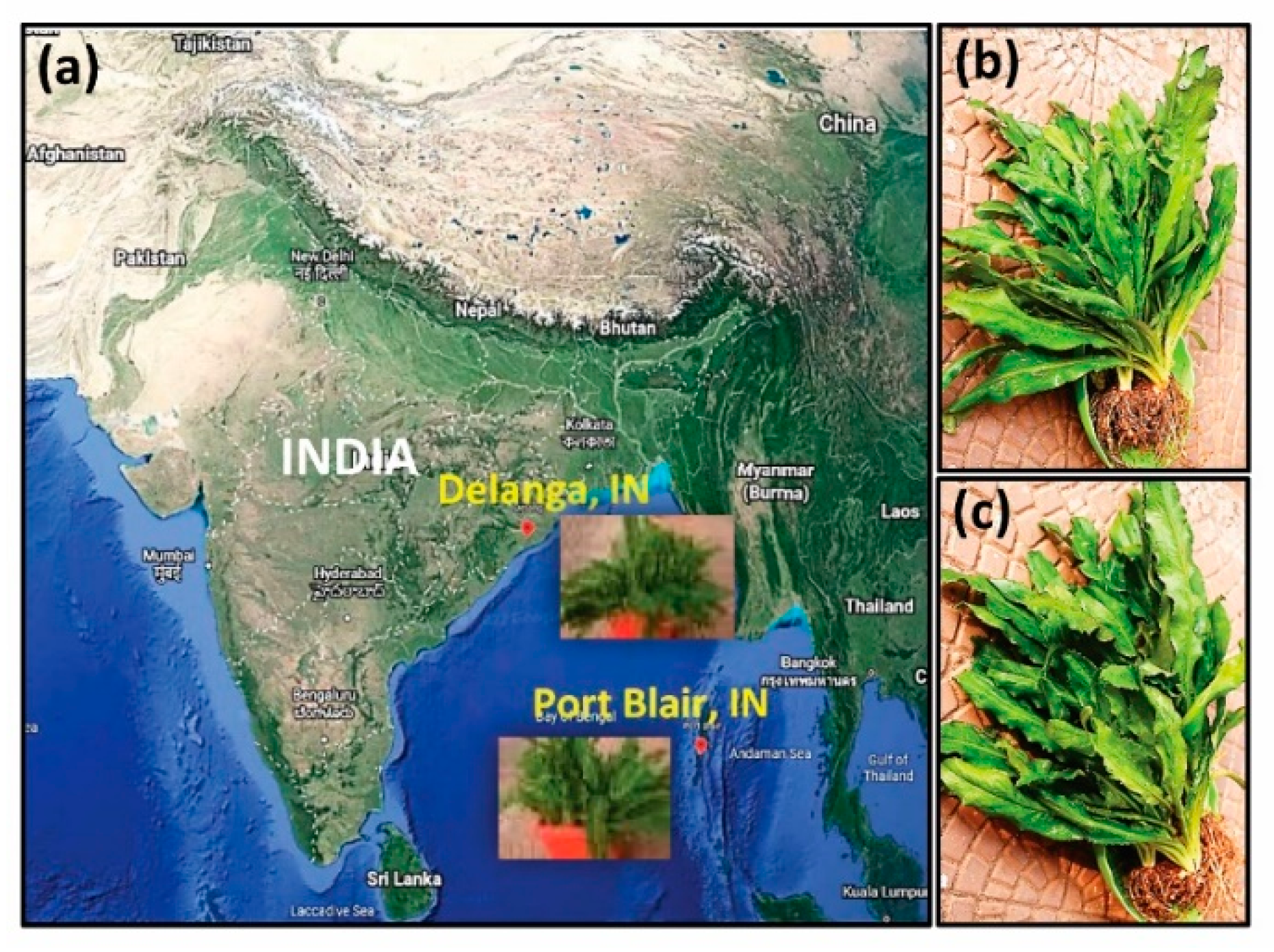
2.1. Plant Materials
2.2. Plant Sample Preparation
2.3. Total Genomic DNA Isolation
2.4. PCR Amplification
2.5. DNA Sequencing
2.6. Bioinformatics Analysis
2.7. DNA Barcoding and ITS2 Secondary Structure Predictions
3. Results
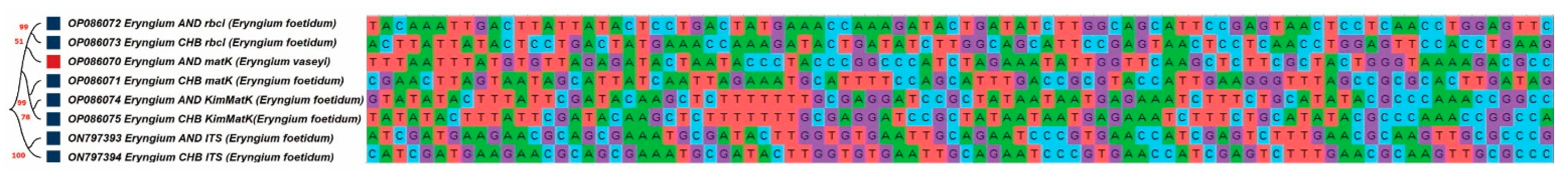
3.1. Amplification, Sequencing, Multiple Sequence Alignment, and Species Identification
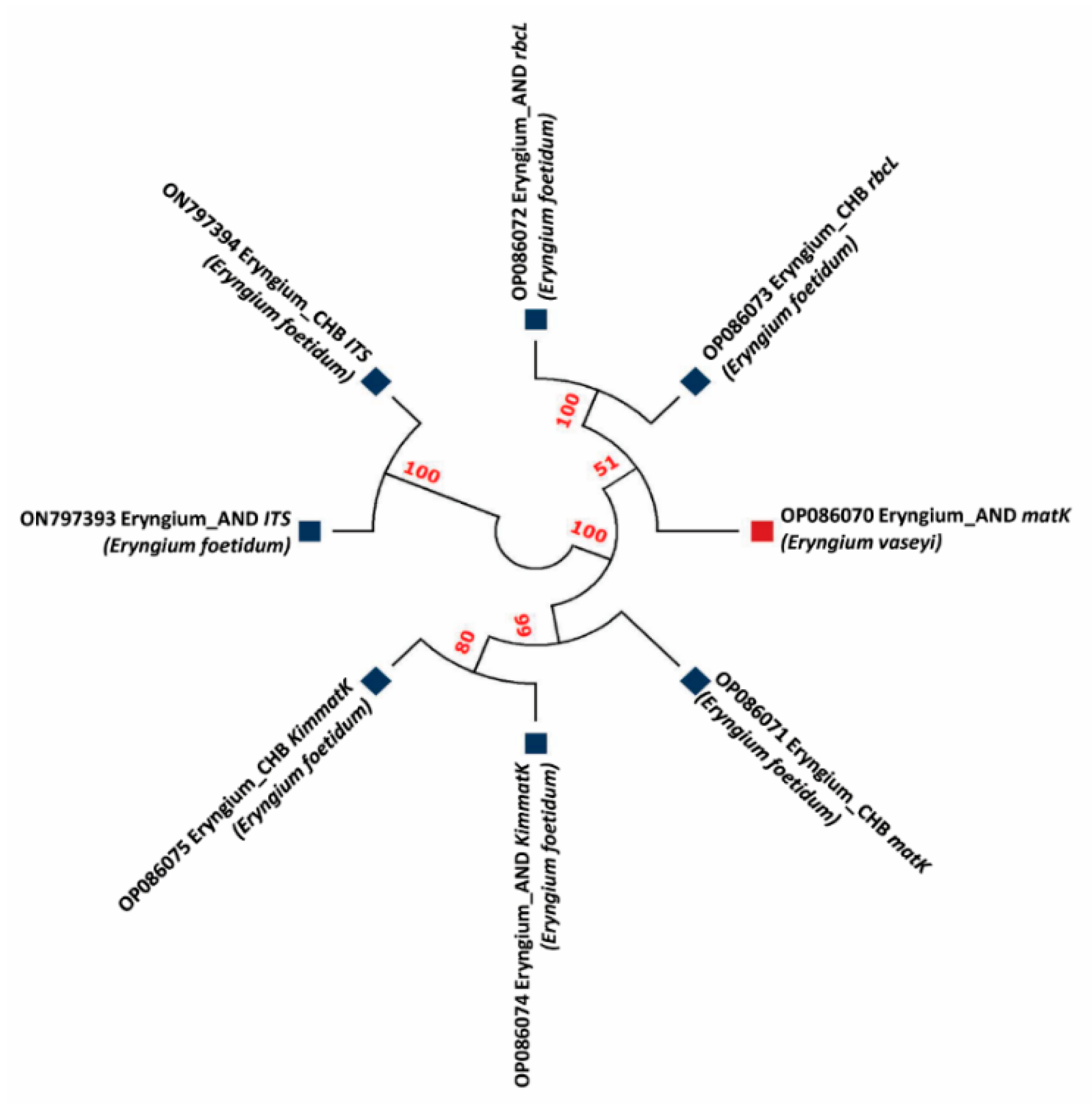
3.2. Phylogenetic Studies
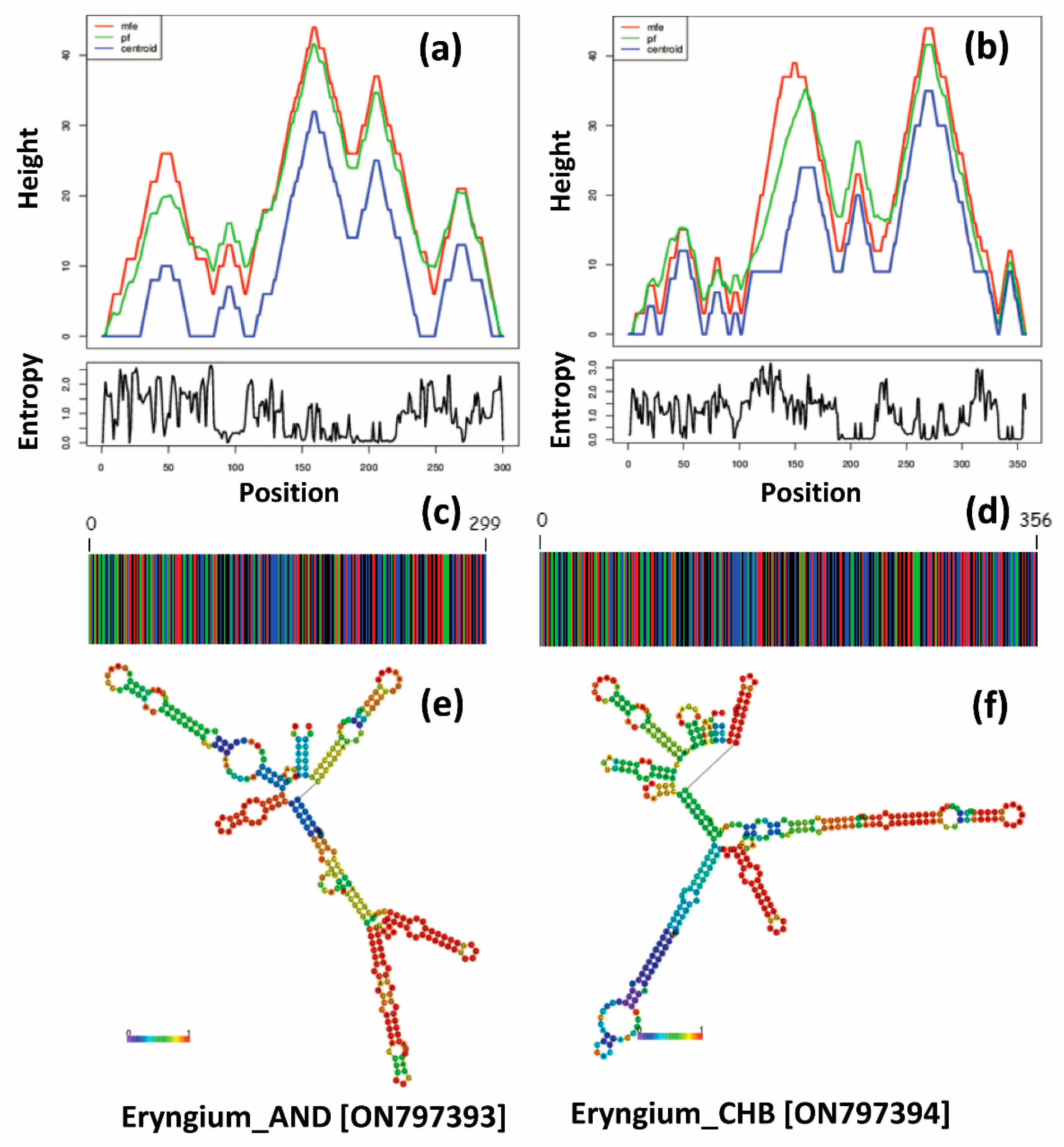
3.3. DNA Barcoding and ITS2 Secondary Structure Predictions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Su, Z.; Yuan, W.; Deng, G.; Li, S. Phytochemical constituents and pharmacological activities of Eryngium L. (Apiaceae). Pharm. Crops 2012, 3, 99–120. [Google Scholar] [CrossRef]
- Le Claire, E.; Schwaiger, S.; Banaigs, B.; Stuppner, H.; Gafner, F. Distribution of a new rosmarinic acid derivative in Eryngium alpinum L. and other Apiaceae. J. Agric. Food Chem. 2005, 53, 4367–4372. [Google Scholar] [CrossRef] [PubMed]
- Ramcharan, C. Culantro: A much utilized, little–understood herb. In Perspectives on New Crops and New Uses; Janick, J., Ed.; ASHS Press: Alexandria, VA, USA, 1999; pp. 506–509. [Google Scholar]
- Singh, B.K.; Ramakrishna, Y.; Ngachan, S.V. Spiny coriander (Eryngium foetidum L.): A commonly used, neglected spicing–culinary herb of Mizoram, India. Genet. Resour. Crop Evol. 2014, 61, 1085–1090. [Google Scholar] [CrossRef]
- Perez-Munoz, E.P.; Antunes-Ricardo, M.; Martínez-Ávila, M.; Guajardo-Flores, D. Eryngium species as a potential ally for treating metabolic syndrome and diabetes. Front. Nutr. 2022, 9, 878306. [Google Scholar] [CrossRef] [PubMed]
- Mekhora, C.; Muangnoi, C.; Chingsuwanrote, P.; Dawilai, S.; Svasti, S.; Chasri, K.; Tuntipopipat, S. Eryngium foetidum suppresses inflammatory mediators produced by macrophages. Asian Pac. J. Cancer Prev. 2012, 13, 723–734. [Google Scholar] [CrossRef]
- Promkum, C.; Butryee, C.; Tuntipopipat, S.; Kupradinun, P. Anticlastogenic effect of Eryngium foetidum L. Assessed by erythrocyte micronucleus assay. Asian Pac. J. Cancer Prev. 2012, 13, 3343–3347. [Google Scholar] [CrossRef]
- Vukic, M.D.; Vukovic, N.L.; Djelic, G.T.; Obradovic, A.; Kacaniova, M.M.; Markovic, S.; Popović, S.; Baskić, D. Phytochemical analysis, antioxidant, antibacterial and cytotoxic activity of different plant organs of Eryngium serbicum L. Ind. Crops Prod. 2018, 115, 88–97. [Google Scholar] [CrossRef]
- Acharya, G.C.; Ponnam, N.; Kumari, M.; Roy, T.K.; Shivashankara, K.S.; Sahoo, M.R. Phytochemical profiling of spiny coriander (Eryngium foetidum L.)—A potential perennial spicing–culinary herb of eastern India. Acta Chromatogr. 2021, 34, 197–202. [Google Scholar] [CrossRef]
- Worz, A. A new subgeneric classification of the genus Eryngium L. (Apiaceae, Saniculoideae). Bot. Jahrb. Syst. 2005, 126, 253–259. [Google Scholar] [CrossRef]
- Calvino, C.I.; Martínez, S.G.; Downie, S.R. The evolutionary history of Eryngium (Apiaceae, Saniculoideae): Rapid radiations, long distance dispersals, and hybridizations. Mol. Phylogenet. Evol. 2007, 46, 1129–1150. [Google Scholar] [CrossRef]
- Jamdade, R.; Mosa, K.A.; El-Keblawy, A.; Al Shaer, K.; Al Harthi, E.; Al Sallani, M.; Al Jasmi, M.; Gairola, S.; Shabana, H.; Mahmoud, T. DNA barcodes for accurate identification of selected medicinal plants (Caryophyllales): Toward barcoding flowering plants of the United Arab Emirates. Diversity 2022, 14, 262. [Google Scholar] [CrossRef]
- Hartvig, I.; Czako, M.; Kjaer, E.D.; Nielsen, L.R.; Theilade, I. The Use of DNA Barcoding in Identification and Conservation of Rosewood (Dalbergia spp.). PLoS ONE 2015, 10, e0138231. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Paulay, G.; Meyer, C.P. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Yu, J.; Xue, J.-H.; Zhou, S.-L. New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Nurhasanah, S.; Papuangan, N. Amplification and analysis of rbcl gene (ribulose-1,5-bisphosphate carboxylase) of clove in ternate Island. IOP Conf. Ser. Earth Environ. Sci. 2019, 276, 012061. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef]
- Bolson, M.; de Camargo Smid, E.; Brotto, M.L.; Silva-Pereira, V. ITS and trnH-psbA as Efficient DNA Barcodes to Identify Threatened Commercial Woody Angiosperms from Southern Brazilian Atlantic Rainforests. PLoS ONE 2015, 10, e0143049. [Google Scholar] [CrossRef]
- Han, S.; Sebastin, R.; Wang, X.; Lee, K.J.; Cho, G.-T.; Hyun, D.Y.; Chung, J.W. Identification of Vicia Species Native to South Korea Using Molecular and Morphological Characteristics. Front. Plant Sci. 2021, 12, 608559. [Google Scholar] [CrossRef]
- Gawronski, P.; Palac, A.; Scharff, L.B. Secondary Structure of Chloroplast mRNAs In Vivo and In Vitro. Plants 2020, 9, 323. [Google Scholar] [CrossRef]
- Yang, X.; Yang, M.; Deng, H.; Ding, Y. New Era of Studying RNA Secondary Structure and Its Influence on Gene Regulation in Plants. Front. Plant Sci. 2018, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. ITS1, 5.8S and ITS2 secondary structure modelling for intra–specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. SpringerPlus 2014, 3, 684. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, A.; Zaman, W.; Saqib, S.; Ullah, F.; Mahmood, T. Phylogeny and diversity of Lamiaceae based on rps14 gene in Pakistan. Genetika 2020, 52, 435–452. [Google Scholar] [CrossRef]
- Al-Juhani, W.S.; Khalik, K.N.A. Identification and phylogenetics study of Arabis alpina L. from the Kingdom of Saudi Arabia. Pak. J. Bot. 2021, 53, 1057–1064. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; HönerzuSiederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithmic Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Zaman, W.; Ye, J.; Ahmad, M.; Saqib, S.; Shinwari, Z.K.; Chen, Z. Phylogenetic exploration of traditional Chinese medicinal plants: A case study on Lamiaceae. Pak. J. Bot. 2022, 54, 1033–1040. [Google Scholar] [CrossRef]
- Mathew, D. DNA barcoding and its application in horticultural crops. In Biotechnology in Horticulture: Methods and Application; Peter, K.K., Ed.; NIPA: New Delhi, India, 2013; pp. 25–50. [Google Scholar] [CrossRef]
- Heise, W.; Barik, W.; Kubosz, D.; Kajtoch, L. A three–marker DNA barcoding approach for ecological studies of xerothermic plants and herbivorous insects from central Europe. Bot. J. Lin. Soc. 2015, 177, 576–592. [Google Scholar] [CrossRef][Green Version]
- Parveen, I.; Singh, H.K.; Malik, S.; Raghuvanshi, S.; Babbar, S.B. Evaluating five different loci (rbcL, rpoB, rpoC1, matK, and ITS) for DNA barcoding of Indian orchids. Genome 2017, 60, 665–671. [Google Scholar] [CrossRef]
- Mathew, D.; Ramesh, G.A. A universal system for matK gene based diagnostic markers to identify the species in Cucurbitaceae. Indian J. Hortic. 2020, 77, 733–735. [Google Scholar] [CrossRef]
- Felsenstein, J. (Ed.) Inferring Phylogenies; Sinauer Associates Inc.: Sunderland, MA, USA, 2004; p. 664. [Google Scholar]
- Zhang, W.; Tian, W.; Gao, Z.; Wang, G.; Zhao, H. Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint. Int. J. Mol. Sci. 2020, 21, 6395. [Google Scholar] [CrossRef] [PubMed]
- Kress, J.W.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- Poovitha, S.; Stalin, N.; Balaji, R.; Parani, M.; Bank, M. Multi-locus DNA barcoding identifies matK as a suitable marker for species identification in Hibiscus L. Genome 2016, 59, 1150–1156. [Google Scholar] [CrossRef]
- Ashfaq, M.; Asif, M.; Anjum, Z.; Zafar, Y. Evaluating the capacity of plant DNA barcodes to discriminate species of cotton (Gossypium: Malvaceae). Mol. Ecol. Res. 2013, 13, 573–582. [Google Scholar] [CrossRef]
- Newmaster, S.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [PubMed]
| Sl. No. | Region | Primer Name | Sequences (5′ to 3′) | Amplicon Size |
|---|---|---|---|---|
| 1 | matK | matK XF | TAATTTACGATCAATTCATTC | 1500 bp |
| matK 5R | GTTCTAGCACAAGAAAGTCG | |||
| 2 | Kim matK | 3F_Kim matK | CGTACAGTACTTTTGTGTTTACGAG | 1500–1580 bp |
| 1R_Kim matK | ACCCAGTCCATCTGGAAATCTTGGTTC | |||
| 3 | rbcL | rbcLa F | ATGTCACCACAAACAGAGACTAAAGC | 600–800 bp |
| rbcLa R | GTAAAATCAAGTCCACCRCG | |||
| 4 | ITS2 | ITS2 S2F | ATGCGATA CTTGGTGTGAATTATAGAAT | 400–800 bp |
| ITS2 S3R | GACGCTTCTCCAGACTACAAT |
| Barcode Genes | Voucher Specimen | Scientific Name | Accession Number | E Value | Query Coverage | Percent Identity |
|---|---|---|---|---|---|---|
| matK | Eryngium_AND | E. vaseyi | OP086070 | 0.0 | 100% | 99.76% |
| Eryngium_CHB | E. foetidum | OP086071 | 0.0 | 100% | 99.81% | |
| Kim matK | Eryngium_AND | E. foetidum | OP086074 | 0.0 | 98% | 99.74% |
| Eryngium_CHB | E. foetidum | OP086075 | 0.0 | 100% | 99.87% | |
| rbcL | Eryngium_AND | E. foetidum | OP086072 | 0.0 | 100% | 100% |
| Eryngium_CHB | E. foetidum | OP086073 | 0.0 | 100% | 100% | |
| ITS2 | Eryngium_AND | E. foetidum | ON797393 | 2 × 10−153 | 100% | 100% |
| Eryngium_CHB | E. foetidum | ON797394 | 2 × 10−153 | 100% | 100% |
| Voucher Specimen | Eryngium_AND | Eryngium_CHB | ||||||
|---|---|---|---|---|---|---|---|---|
| Markers | matK | Kim matK | rbcL | ITS2 | matK | Kim matK | rbcL | ITS2 |
| Sequence length | 823 | 763 | 493 | 300 | 534 | 780 | 476 | 357 |
| Alignment length | 823 | 763 | 492 | 300 | 533 | 773 | 419 | 357 |
| Maximum score | 1509 | 1399 | 909 | 555 | 979 | 1424 | 774 | 660 |
| GC content (%) | 37.18 | 36.70 | 43.00 | 62.33 | 35.96 | 36.28 | 43.28 | 63.31 |
| CDS region | 2 | 1 | 1 | – | 2 | 1 | 1 | – |
| Amino acid | 274 | 254 | 164 | – | 177 | 260 | 159 | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acharya, G.C.; Mohanty, S.; Dasgupta, M.; Sahu, S.; Singh, S.; Koundinya, A.V.V.; Kumari, M.; Naresh, P.; Sahoo, M.R. Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India. Genes 2022, 13, 1678. https://doi.org/10.3390/genes13091678
Acharya GC, Mohanty S, Dasgupta M, Sahu S, Singh S, Koundinya AVV, Kumari M, Naresh P, Sahoo MR. Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India. Genes. 2022; 13(9):1678. https://doi.org/10.3390/genes13091678
Chicago/Turabian StyleAcharya, Gobinda Chandra, Sansuta Mohanty, Madhumita Dasgupta, Supriya Sahu, Satyapriya Singh, Ayyagari V. V. Koundinya, Meenu Kumari, Ponnam Naresh, and Manas Ranjan Sahoo. 2022. "Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India" Genes 13, no. 9: 1678. https://doi.org/10.3390/genes13091678
APA StyleAcharya, G. C., Mohanty, S., Dasgupta, M., Sahu, S., Singh, S., Koundinya, A. V. V., Kumari, M., Naresh, P., & Sahoo, M. R. (2022). Molecular Phylogeny, DNA Barcoding, and ITS2 Secondary Structure Predictions in the Medicinally Important Eryngium Genotypes of East Coast Region of India. Genes, 13(9), 1678. https://doi.org/10.3390/genes13091678









