Abstract
Background: There are few studies on the burden of chromosomal abnormalities and single gene disorders in fetal hemivertebra (HV). We aim to investigate the cytogenetic and monogenic risk and evaluate prenatal outcomes of fetal HV. Method: This study included fetuses diagnosed with HV divided into two groups: isolated HV and non-isolated HV. Data on other sonographic structural anomalies, chromosomal and sub-chromosomal abnormalities, monogenic variations detected by WES, and prenatal outcomes are recorded and reviewed. Results: Among 109 fetal HV cases, forty-seven (43.1%) non-isolated HV cases were associated with structural anomalies. Chromosomal test results were available in 58 cases, identifying six (10.3%) chromosomal aberrations involved in four isolated and two non-isolated HV. WES identified four (likely) pathogenic variants in three cases among 16 fetuses with HV, involving three novel variants, 1250G > T and c.1277G> inherited from parents, respectively, in DLL3 and c.7213C > A ** in the FLNB. The live birth rate (LB) was higher in the isolated fetal HV group than in the non-isolated group (67.7% (42/62) vs. 12.5% (12/47), p < 0.001). Conclusion: This study emphasizes the risk of cytogenetic abnormalities in isolated HV. WES yields a diagnostic rate of 18.3% in HV with normal CMA, probably aiding the prenatal counseling and management of fetal HV.
1. Introduction
Hemivertebra is the most frequent vertebral anomaly of congenital spinal abnormalities, such as congenital scoliosis and kyphosis, with an incidence of about 0.1–1.0% in live births [1]. The patients with HV deformity tend to experience spinal cord strain due to a loss of normal physiologic curvature of the spine. The HV may affect the development and function of the heart, lungs, and other vital organs by compression. Other associated structural malformations are observed in about 59.2–71.2% of fetuses with HV involving the cardiovascular system, genitourinary system, skeletal system, and central nervous system, etc. [1,2,3]. Moreover, 25.9% of children diagnosed prenatally with HV experience developmental delay [2]. Possible causes of fetal HV include failure of chondrification, spinal formation and development, and nutritional deficiency resulting from abnormal distribution of nutrients [4,5]. Despite this, few studies have been conducted on chromosomal abnormalities and single gene disorders in fetuses with prenatal HV.
As chromosomal microarrays (CMA) and whole exome sequence (WES) have been rapidly developed and applied to prenatal diagnosis, CMA improves the diagnostic rate by 6–10% [6,7,8], and WES yields an additional diagnostic rate of approximately 10% in fetal sonographic structural abnormalities, respectively [9]. Although a few small-sample studies suggest that chromosomal abnormalities are unlikely in isolated fetal HV, there is still debate in clinical practice about how to use and apply cytogenetic tools for fetal HV due to the lack of evidence. In a systematic review, 326 fetal HV were found to have 5% cytogenetic abnormalities, including partial tetrasomy 4q, mosaic trisomy 4, mosaic trisomy 7, mosaic trisomy 9, mosaic trisomy 18q, mosaic trisomy 18; trisomy 7, trisomy 15q with monosomy 6q, partial trisomy 22; duplication of 2p; 4p- deletion, 17p deletion, 18p deletion, 18q22.2 deletion, 22q13.3 deletion; ring chromosome 21, and Fanconi’s anemia [10]. Furthermore, HV is linked with numerous genetic disorders, including DLL3, MESP2, LFNG, HES7, TBX6, RIPPLY2, GDF6, and EBP genes, etc. [11]. However, rather than exome sequencing, most of the studies on the genetic causes of HV are typically conducted using gene panels or particular gene detection techniques. WES is still understudied and underutilized when it comes to fetal HV, causing confusion and challenges in genetic counseling and management for pregnancies with fetal HV in the prenatal setting.
This study investigates chromosomal and sub-chromosomal abnormalities as well as monogenic conditions in fetal HV and assesses the efficacy and performance of both CMA and WES in pregnancy. In addition, the perinatal and postnatal prognosis of fetal HV is evaluated.
2. Materials and Methods
A retrospective study was conducted to identify the cases with fetal hemivertebra identified by prenatal ultrasound or nuclear magnetic resonance imaging (MRI) from 2015 to 2021 in the Guangzhou Women and Children’s Medical Center. This study was approved by the Ethics Committee of the Guangzhou Women and Children’s Medical Center. The HV is diagnosed if the strong echoes of triangular and irregular solitary bony structures are found within the vertebral columns, and the two strong echo lines of the spine are not parallel. Parents were offered chromosomal microarray analysis (CMA) after completing pretest counseling and obtaining informed consent. Demographic and genetic data were collected, including maternal age, gestational age at which HV suspicion arose, stage of the disease, other structural abnormalities associated with HV, and chromosomal and genetic test results of the invasive procedures and perinatal outcomes, etc. The acronym VACTERL is defined by the presence of at least three of the following: Vertebral, Anal, Cardiac, Tracheoesophageal fistula or Esophageal atresia, Renal, and Limb anywhere. The information on postnatal outcomes was obtained from the medical record system and telephone follow-up. Preterm birth was defined as a gestational age < 37 weeks after gestational age was assessed by maternal menstrual history and ultrasound in the first trimester. The pediatric medical records were reviewed to investigate the onset of scoliosis or other spinal abnormalities and whether an operation was needed.
The DNA sample was extracted by chorionic villus cells, amniocytes, umbilical cord blood cells, and peripheral blood lymphocytes by using a QIAamp DNA Blood Midi/Mini kit (QIAGEN GmbH; Hilden, Germany). The quantitative fluorescent polymerase chain reaction (QF-PCR) was used to exclude maternal cell contamination and abnormalities of chromosomes 21, 18, 13, and sex chromosomes. The samples were subjected to CMA when there were normal QF-PCR results. CMA was conducted by using Affymetrix CytoScan HD/750K array with a single-nucleotide polymorphism array (SNP array) and array-based comparative genomic hybridization (aCGH) platforms at resolutions of 10 and 100 kb, respectively, according to the manufacturer’s protocol (Affymetrix Inc., Santa Clara, CA, USA) as in our previous study [12]. The genome built was referred to as GRCh37/hg19. The classification of the copy number variants was according to joint consensus recommendations of the American College of Medical Genetics and Genomics and ClinGen [13]. The pathogenic CNVs, likely pathogenic CNVs, and variants of unknown significance (VUS) are recorded and documented, but likely benign and benign VUS are not considered. Parental CMA is advised for couples to determine the source of CNVs when fetal CNVs are discovered. The samples were subjected to Trio-WES once there were normal QF-PCR and CMA results. According to the manufacturer’s protocol, the DNA samples were enriched using Agilent SureSelect human exome capture probes (V6, Life Technologies, Carlsbad, CA, USA). The pair-end 150-bp reads’ libraries were sequenced using Hiseq XTen (Illumina, Inc., San Diego, CA, USA). NextGENe v2.4.1.2 software (SoftGenetics, State College, PA, USA) was used for variants calling. After filtering out the synonymous and common SNPs (MAF > 0.1%), rare variants with high confidence were considered disease-causing candidates. Variant annotation was further confirmed through literature and population databases, including 1000 Genomes, dbSNP, GnomAD, Clinvar, HGMD, and OMIM. Multiple computational algorithms, including SIFT, MutationTaster, PolyPhen2, PROVEAN, CADD, Human Splicing Finder, MaxEntScan, and NNSplice, were used to evaluate the pathogenicity of the candidate gene variants. The interpretation of the variants was performed according to the American College of Medical Genetics’ guidelines [14].
Statistical analysis was performed using the IBM statistical program SPSS 25.0. The mean and range were used to describe the demographical characteristics. The Chi-square test was used for categorical data in appropriate cases. A p-value < 0.05 was considered statistically significant.
3. Results
A total of 109 cases with prenatal HV were included in the study flowchart shown in Figure 1. There were 58 fetuses with chromosomal test results from invasive procedures with six (10.3%) diagnostic pathogenic copy number variants. The whole exome sequence yielded a detection rate of 18.8% in 16 fetuses with HV. The outcome included 51 terminations of pregnancy, four lost follow-ups, and 54 live births, including 42 full-term births and 12 preterm births. Nine (33.3%) of the 27 terminations of pregnancies with the available results of the invasive procedures used the prenatal genetic diagnosis information to assist them in making the choice to terminate the pregnancy, seven cases due to the diagnosis of other malformations, and the voluntary abortion in the remaining 11 cases. The live birth rate (LB) was higher in the isolated fetal HV group than in the non-isolated group (67.7% (42/62) vs. 12.5% (12/47), p < 0.001). Preterm births occurred in 19.4% (8/42) of isolated HV group and 33.3% (4/12) of non-isolated group (p = 0.431). Among the live birth cases, there was an operation rate of 28.6% (12/42) in the isolated group and 58.3% (7/12) in the non-isolated group (p = 0.087). The pathogenic or likely pathogenic variants identified on CMA or WES results only, and the clinical outcomes of fetal HV are presented in Table 1. The details of genetic and clinical outcomes of fetal HV are presented in Table 1. Spinal surgery was required in 19 cases. The mean follow-up period was 2.3 years (range 0.2–8.1 years). All of the live birth cases were found to be without developmental delay except that one female case with HV combined with a butterfly vertebra, who had normal CMA, and later developed seizures postnatally.
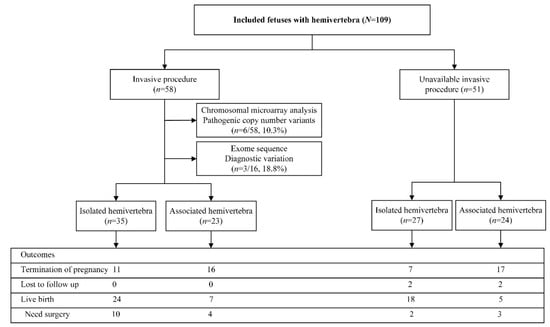
Figure 1.
Flowchart of this study.

Table 1.
Genetic and clinical outcome in fetuses identified sonographically in fetuses with hemivertebra.
The mean maternal age was 30.7 years (range 19.0–42.0 years), and the mean gestational age at suspected HV was 23.9 weeks (range 15+1–32+0 weeks). Among 62 (56.9%) cases with isolated HV, 44 fetuses had a single HV at the cervical (n = 3), thoracic (n = 24), lumbar (n = 16), and sacral (n = 1) vertebras. Eighteen fetuses were found with multiple stages of vertebral involvement. Another 47 (43.1%) non-isolated HV cases were associated with structural anomalies. Table 2 shows the details of associated anomalies identified sonographically in fetuses with HV. The skeletal system (19.1%) and genitourinary system anomalies (19.1%) were the most frequently associated structural anomalies, followed by the cardiovascular system (16.2%), central nervous system (10.3%), craniofacial abnormalities (8.8%) and gastrointestinal system (5.9%). Other structural anomalies included polyhydramnios, fetal growth restriction, oligohydramnios, and pleural effusion. In addition, VACTERL syndrome was identified in three cases.

Table 2.
Associated anomalies identified sonographically in fetuses with hemivertebra.
CMA detected six (10.3%) clinically significant variants (pCNVs) and five (8.6%) variants of uncertain significance (VOUS). The clinically significant variants included mosaic trisomy 21, maternal uniparental disomy for chromosome 15, 16p11.2 microdeletion, 17p11.2 microdeletion, 8q24.3 microduplication, and 21q22.2q22.3 microdeletion involved in four fetuses with isolated HV and two with non-isolated HV (Table 3). Parental CMA results were available in five cases with P/LP or VUS, but the remaining patients refused the test because it costs more than USD1000. Among the five cases, the CNVs identified by CMA were de novo in patients 1, 2, 3, and 6. However, the VUS detected in patient 7 was inherited from his father who did not exhibit associated phenotypes. The 32 cases with negative CMA did not undergo WES mainly because the patients rejected trio-WES due to its high cost, which can reach USD1500 in China.

Table 3.
Pathogenic copy number variation and VUS identified by CMA in prenatal hemivertebra cases.
As shown in Table 4, the whole exome sequence identified four (likely) pathogenic variants in three genes and three variants of uncertain significance (VUS) in normal CMA cases. The pregnancy of patient 12 with a two-times birth history of spinal anomalies, identified with multiple stages of HV and multi-segment fusion, was detected with two novel and missense mutations, NM_203486.3, c.1250G > T (p. Cys417Phe) and c.1277G > A (p. Cys426Tyr) inherited from the father and mother, respectively, in the DLL3 gene, with autosomal recessive inheritance mode, leading to spondylocostal dysostosis 1 (OMIM: 277300). In patient 13, the fetus was found with thoracic HV and short limb long bone (<−3SD) and identified with a de novo heterozygous variant in the EBP gene, NM_006579.2, c.328C > T (p. Arg110Ter) with AD mode, which could result in X-linked dominant chondrodysplasia punctata and MEND syndrome (OMIM: 302960, 300960). Patient 14 was diagnosed with thoracic HV associated with sacrococcygeal spina bifida. In this fetus, WES detected a mutation in the FLNB gene, NM_001164317.1, c.7213C > A ** (p. Arg2405Ser). This variant was de novo, pathogenic, and nonsense and associated with Larsen syndrome (OMIM: 150250).

Table 4.
Diagnostic variants detected by whole exome sequence in fetuses with hemivertebra.
4. Discussion
Hemivertebra is the most common cause of congenital vertebral abnormalities, with an approximate incidence of 0.1–1.0% of births [1]. With the wide application and use of chromosomal microarray analysis and whole exome sequence in the prenatal setting, chromosomal, sub-chromosomal, and genetic etiologies of prenatal skeletal abnormalities have been well studied and understood [6,15]. However, little research has been conducted on the cytogenetic and monogenic burden and outcomes in fetuses with HV. The study aimed to evaluate the efficacy of CMA and WES, and the perinatal and postnatal outcomes in fetal HV.
There was a high prevalence of associated structural anomalies in prenatal fetuses with hemivertebra. Previous studies found co-existing abnormalities in 59.2–71.2% of fetal HV cases [1,2,3]. This study found 43.1% of cases diagnosed with other structural defects, lower than the previous data. As a tertiary referral center, there was likely a selective referral bias, and more minute and minor changes in fetal vertebral columns could be identified by the increasingly better resolution of the ultrasound and magnetic resonance machine. Furthermore, similar to previous studies [4,11], in these fetuses with associated anomalies, the incidence was high in the genitourinary, skeletal, and cardiovascular system, followed by central nervous systems, etc. Additionally, it was important to note that there were three HV fetuses with fetal growth restriction. A recent study proved the association between the supernumerary hemivertebra and intrauterine growth restriction [16]. Thus, this suggested that it was necessary to conduct a thorough anomaly scan, biometry monitoring and echocardiogram for fetal HV.
It seems that chromosomal abnormalities are not usually associated with isolated fetal HV [1,2,16]. In addition, a recent systematic review of prenatal cytogenetics of the fetal hemivertebra showed that cytogenetic abnormalities accounted for 5% of 246 fetal HV cases, and what is noteworthy is that all of the cases had prenatally (29.0%) or postnatally (71.0%) associated anomalies [10]. However, isolated fetal HV was indeed associated with chromosomal and sub-chromosomal abnormalities. In our series, it was noteworthy that there were four fetuses with isolated HV and two with non-isolated HV among all six with cytogenetic diagnoses. It implied that chromosomal abnormalities could be present in fetuses with isolated HV. Because some co-existing anomalies may have been missed by prenatal ultrasound and likely observed postnatally, the so-called “prenatal isolated HV” may not actually represent true isolated HV. In short, a diagnostic yield of 10.3% warrants the application of CMA in pregnancies with HV.
Congenital spinal abnormalities can result from the 16p11.2 microdeletion syndrome [17]. We found two cases of 16p11.2 microdeletion syndrome in patients 5 and 6 with isolated HV. According to a postnatal study of congenital scoliosis, 10% of affected cases could be explained by the 16p11.2 microdeletion [18]. Additionally, a study of prenatal sonographic features in 16p11.2 microdeletion syndrome found that 41.7% of fetuses had spinal defects, including HV and butterfly vertebra [19]. The TBX6 gene, which is located on the 16p11.2 chromosome, has been shown through animal studies to be crucial in congenital spinal abnormalities. The primary mechanism involves the haploinsufficiency mechanism, which leads to reduced regulation of downstream genes due to mislocalization of the T-Box transcription factor 6, thereby dysregulating the Notch signaling pathway, which is essential for somite development [20,21,22,23]. However, it is crucial to note that increased doses of TBX6 are responsible for congenital malformations of cervical vertebrae [24]. This suggests that precise dosages of the TBX6 gene have a crucial impact on spinal development. Furthermore, the hemivertebra and scoliosis resulted from the nonfunctional allele in combination with another common hypomorphic allele haplotype T-C-A (three common single nucleotide polymorphisms: rs2289292, rs3809624, rs3809627) in TBX6 [25]. The frequency of the T-C-A haplotype reaches 44% among Han Chinese individuals in the 1000 Genomes Project [26]. Therefore, the allelic haplotype T-C-A of TBX6 should not be overlooked in hemivertebra and congenital scoliosis.
Previous research indicated that the whole exome sequence could provide approximately 8.5–10.0% diagnostic yield in fetal structural anomalies detected by ultrasound in normal CMA [9,27]. While the application and utility of WES in fetal HV was still unclear, our data showed that WES improved an 18.8% genetic diagnostic rate for fetal HV over CMA. All of the three cases with diagnostic variants were detected with multiple HV, and two of them had short limb bone and spinal bifida, respectively. Furthermore, the whole exome sequence can identify additional novel variants and findings. This was the first report of the association between fetal multiple HV and DLL3, demonstrating the prenatal phenotype for DLL3. Two novel variants in the DLL3 gene, transmitted from the parents, were detected in patient 12 with multiple HV, c.1250G > T (p. Cys417Phe), and c.1277G > A (p. Cys426Tyr). In addition, it suggested that each sibling of an affected individual had a 25% chance of being affected by the two novel variants of the DLL3 gene in this family. In the genetic counseling of this couple, there were at least two options. First, the chorionic villus sampling is performed to exclude the two variants of the DLL3 gene in a subsequent pregnancy as early as in the first trimester. Secondly, pre-implantation for monogenic disease (PGT-M) can be a selection to rule out the two variants of the DLL3 gene during the in vivo fertilization (IVF) process. Additionally, a novel mutation in the FLNB gene, c.7213C > A ** (p. Arg2405Ser), was identified in patient 14. This variant was de novo and absent from the parents, meaning that the recurrent risk of this variant would be no more than 3% in the following pregnancy, except for the parental germline mosaicism. Therefore, if fetuses with HV have been identified by ultrasound or MRI, particularly when multiple segmentation of HV or co-existing structural abnormalities have been diagnosed, whole exome sequencing could be offered to provide information on genetic variants for cases with normal CMA.
The isolated HV group had better prenatal and postnatal outcomes than the non-isolated group. Our data showed higher birth rates in isolated prenatal cases with hemivertebra. Similar to previous investigations, fetal survival can be lowered if additional structural defects are found [3]. Additionally, preterm rates and surgery rates appeared to be lower in isolated HV cases compared to hemivertebra, despite there being no significant difference. Surprisingly, 53 live births achieved good results with a mean follow-up of 2.3 years, and no developmental delay was found except in one patient with epilepsy. Long-term follow-up care is required. Prenatal CMA was normal in 31 out of the 54 birth cases, including 13 cases that were undiagnosed through WES. It is intriguing to see if patients with normal cytogenetic testing and those without prenatal genetic testing have different long-term follow-up prognosis. To sum up, our study supported the favorable outcome in fetuses with isolated HV diagnosed with normal CMA or WES.
There are several limitations in this study. The recall bias may be present in this retrospective study. Prenatal skeletal abnormalities can be associated with methylation abnormalities and deep intron variations not detectable by CMA or WES. This is a small-sample study, and more research and evidence are needed to investigate the genetic burden of fetal HV.
5. Conclusions
In conclusion, we confirmed the high prevalence of co-existing anomalies in fetuses with hemivertebra. This study showed that the risk of chromosomal and sub-chromosomal abnormalities identified by CMA was as high as 10.3% for fetal hemivertebra with and without other structural defects. WES can yield an 18.8% diagnostic rate in fetuses with normal CMA for fetal hemivertebra, particularly when multiple vertebras are affected and accompanied by other structural abnormalities. Achieving prenatal chromosomal and genetic diagnosis by CMA and WES in fetal hemivertebra provides further information for management and couple counselling.
Author Contributions
Conceptualization and writing: H.Z., Y.W., R.H. and C.L.; Chromosomal microarray data analysis: R.L., X.J. and Y.Z. Exome sequence variants classification: F.F., R.L. and D.W.; Bioinformatics analysis: Q.Y.; Clinical data statistics: K.C., T.L., J.H., X.Y. and D.L.; Supervision and editing: C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the sub-project of the National Key Research and Developmental Program (2021YFC2701002), the National Natural Science Foundation of China (81801461, 81873836, 81771594, 81671474, 81501267), the Natural Science Foundation of Guangdong Province (2019A1515012034, 2017A030313460), and The Project of Guangzhou Science and Technology, Grant/Award Number 202102020191.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Guangzhou Women and Children’s Medical Center with approval code [2018]021402.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
We appreciate all of the participants in this study. We thank all the technical staff from Beijing DE yi Oriental Translational Medicine Research Center for their tremendous technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Basude, S.; McDermott, L.; Newell, S.; Wreyford, B.; Denbow, M.; Hutchinson, J.; Abdel-Fattah, S. Fetal hemivertebra: Associations and perinatal outcome. Ultrasound Obstet. Gynecol. 2015, 45, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Lemire, G.T.; Beauregard-Lacroix, É.; Campeau, P.M.; Parent, S.; Roy-Beaudry, M.; Soglio, D.D.; Grignon, A.; Rypens, F.; Wavrant, S.; Laberge, A.-M.; et al. Retrospective analysis of fetal vertebral defects: Associated anomalies, etiologies, and outcome. Am. J. Med. Genet. Part A 2020, 182, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Zelop, C.M.; Pretorius, D.H.; Benacerraf, B.R. Fetal hemivertebrae: Associated anomalies, significance, and outcome. Obstet. Gynecol. 1993, 81, 412–416. [Google Scholar] [PubMed]
- Johal, J.; Loukas, M.; Fisahn, C.; Chapman, J.R.; Oskouian, R.J.; Tubbs, R.S. Hemivertebrae: A comprehensive review of embryology, imaging, classification, and management. Child’s Nerv. Syst. 2016, 32, 2105–2109. [Google Scholar] [CrossRef]
- Tanaka, T.; Uhthoff, H.K. The Pathogenesis of Congenital Vertebral Malformations: A Study Based on Observations made in 11 Human Embryos and Fetuses. Acta Orthop. Scand. 1981, 52, 413–425. [Google Scholar] [CrossRef]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal Microarray versus Karyotyping for Prenatal Diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef]
- Levy, B.; Wapner, R. Prenatal diagnosis by chromosomal microarray analysis. Fertil. Steril. 2018, 109, 201–212. [Google Scholar] [CrossRef]
- Qi, Q.; Jiang, Y.; Zhou, X.; Meng, H.; Hao, N.; Chang, J.; Bai, J.; Wang, C.; Wang, M.; Guo, J.; et al. Simultaneous Detection of CNVs and SNVs Improves the Diagnostic Yield of Fetuses with Ultrasound Anomalies and Normal Karyotypes. Genes 2020, 11, 1397. [Google Scholar] [CrossRef]
- Petrovski, S.; Aggarwal, V.; Giordano, J.L.; Stosic, M.; Wou, K.; Bier, L.; Spiegel, E.; Brennan, K.; Stong, N.; Jobanputra, V.; et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: A prospective cohort study. Lancet 2019, 393, 758–767. [Google Scholar] [CrossRef]
- Powel, J.E.; Spiliopoulos, M.; Ferreira, C.R.; Rosenthal, E.; Sinkovskaya, E.; Brown, S.; Sham, C.; Jelin, A.C.; Al Kouatly, H.B. 265 Hemivertebra: Systematic review of cases with cytogenetic abnormalities, associated anomalies and proposed prenatal management. Am. J. Obstet. Gynecol. 2021, 224, S175. [Google Scholar] [CrossRef]
- Powel, J.E.; Spiliopoulos, M.; Ferreira, C.R.; Rosenthal, E.; Sinkovskaya, E.; Brown, S.; Sham, C.; Jelin, A.C.; Al Kouatly, H.B. 346 Review of syndromic genetic etiology in patients with hemivertebra and implications in prenatal management. Am. J. Obstet. Gynecol. 2021, 224, S227–S228. [Google Scholar] [CrossRef]
- Zhou, H.; Cheng, K.; Li, Y.; Fu, F.; Li, R.; Zhang, Y.; Yang, X.; Jing, X.; Li, F.; Han, J.; et al. The Genetic and Clinical Outcomes in Fetuses with Isolated Fetal Growth Restriction: A Chinese Single-Center Retrospective Study. Front Genet. 2022, 13, 856522. [Google Scholar] [CrossRef] [PubMed]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257, Correction in: Genet. Med. 2021, 23, 2230. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Pauta, M.; Martinez-Portilla, R.J.; Borrell, A. Diagnostic yield of exome sequencing in fetuses with multisystem malformations: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2022, 59, 715–722. [Google Scholar] [CrossRef]
- Bohiltea, R.E.; Ducu, I.; Mihai, B.M.; Iordache, A.-M.; Dima, V.; Vladareanu, E.M.; Bacalbasa, N.; Bohiltea, A.-T.; Salmen, T.; Varlas, V. First-Trimester Diagnosis of Supernumerary Hemivertebra. Diagnostics 2022, 12, 373. [Google Scholar] [CrossRef]
- Yue, F.; Xi, Q.; Zhang, X.; Jiang, Y.; Zhang, H.; Liu, R. Molecular cytogenetic characterization of 16p11.2 microdeletions with diverse prenatal phenotypes: Four cases report and literature review. Taiwan. J. Obstet. Gynecol. 2022, 61, 544–550. [Google Scholar] [CrossRef]
- Lai, W.; Feng, X.; Yue, M.; Cheung, P.; Choi, V.; Song, Y.-Q.; Luk, K.; Cheung, J.; Gao, B. Identification of Copy Number Variants in a Southern Chinese Cohort of Patients with Congenital Scoliosis. Genes 2021, 12, 1213. [Google Scholar] [CrossRef]
- Lin, S.; Shi, S.; Zhou, Y.; Ji, Y.; Huang, P.; Wu, J.; Chen, B.; Luo, Y. Intrauterine phenotypic features associated with 16p11.2 recurrent microdeletions. Prenat. Diagn. 2018, 38, 381–389. [Google Scholar] [CrossRef]
- White, P.H.; Farkas, D.R.; McFadden, E.E.; Chapman, D.L. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development 2003, 130, 1681–1690. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, M.; Kawakami, A.; Sawada, A.; Furutani-Seiki, M.; Takeda, H.; Araki, K. Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat. Genet. 2002, 31, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, D.B.; McInerney-Leo, A.; Gucev, Z.S.; Gardiner, B.; Marshall, M.; Leo, P.J.; Chapman, D.L.; Tasic, V.; Shishko, A.; Brown, M.A.; et al. Autosomal dominant spondylocostal dysostosis is caused by mutation in TBX6. Hum. Mol. Genet. 2013, 22, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, M.; Duffourd, Y.; Jouan, T.; Poe, C.; Jean-Marçais, N.; Verloes, A.; St-Onge, J.; Riviere, J.-B.; Petit, F.; Pierquin, G.; et al. Autosomal recessive variations of TBX6, from congenital scoliosis to spondylocostal dysostosis. Clin. Genet. 2016, 91, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yang, N.; Wu, N.; Xu, X.; Chen, W.; Zhang, L.; Li, Y.; Du, R.-Q.; Dong, S.; Zhao, S.; et al. Increased TBX6 gene dosages induce congenital cervical vertebral malformations in humans and mice. J. Med. Genet. 2020, 57, 371–379. [Google Scholar] [CrossRef]
- Wu, N.; Ming, X.; Xiao, J.; Wu, Z.; Chen, X.; Shinawi, M.; Shen, Y.; Yu, G.; Liu, J.; Xie, H.; et al. TBX6 Null Variants and a Common Hypomorphic Allele in Congenital Scoliosis. N. Engl. J. Med. 2015, 372, 341–350. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Lord, J.; McMullan, D.J.; Eberhardt, R.Y.; Rinck, G.; Hamilton, S.J.; Quinlan-Jones, E.; Prigmore, E.; Keelagher, R.; Best, S.K.; Carey, G.K.; et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): A cohort study. Lancet 2019, 393, 747–757. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).