SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Preparation
2.2. Neonatal Rat Cardiomyocyte (NRCMs) Isolation, Culture, and Treatment
2.3. Plasmid Constructs
2.4. RT-PCR Analysis
2.5. Protein Expression and Purification from BL21(DE3)
2.6. In Vitro Kinase Assays
2.7. Middle-Down Mass Spectrometry (MS)
2.8. Western Blot
2.9. Co-Immunoprecipitation (co-IP)
2.10. Data Analysis
3. Results
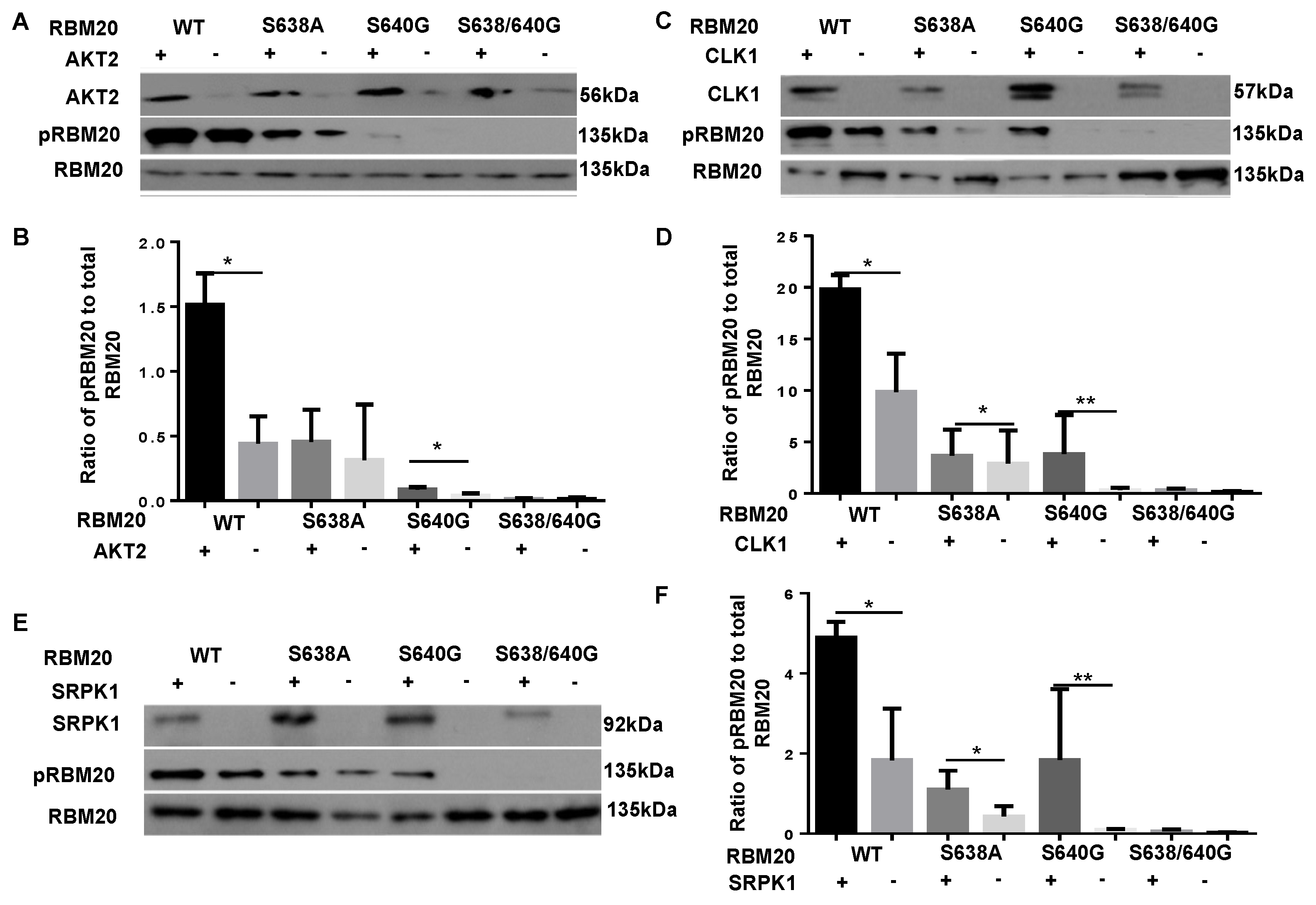
3.1. Co-Transfection with AKT2, CLK1, or SRPK1 Increases Phosphorylation of the RBM20 RSRSP Stretch in HeLa Cells
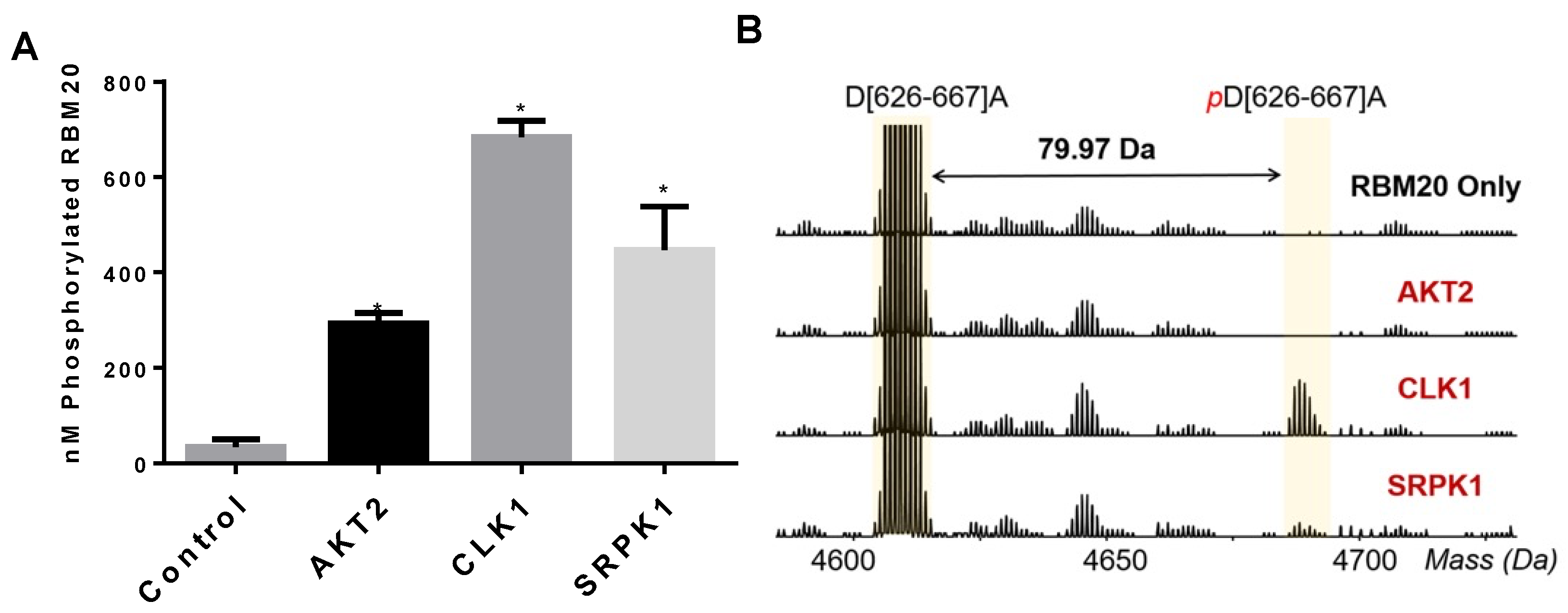
3.2. CLK1 and SRPK1 Directly Phosphorylate the RBM20 RS Domain In Vitro
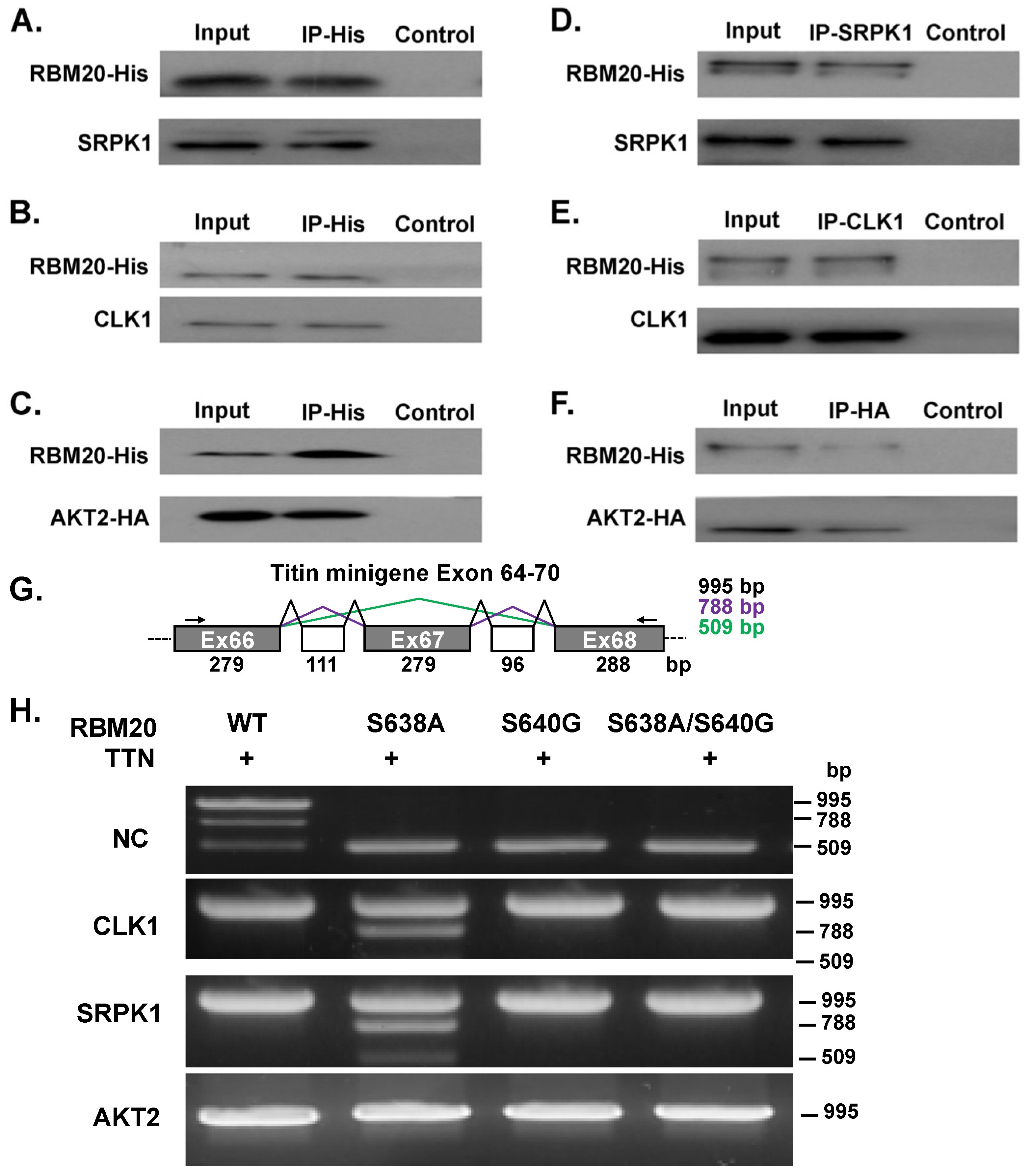
3.3. AKT2, CLK1, and SRPK1 Interact with RBM20 in Co-Transfected HeLa Cells and Regulate Titin Pre-mRNA Splicing
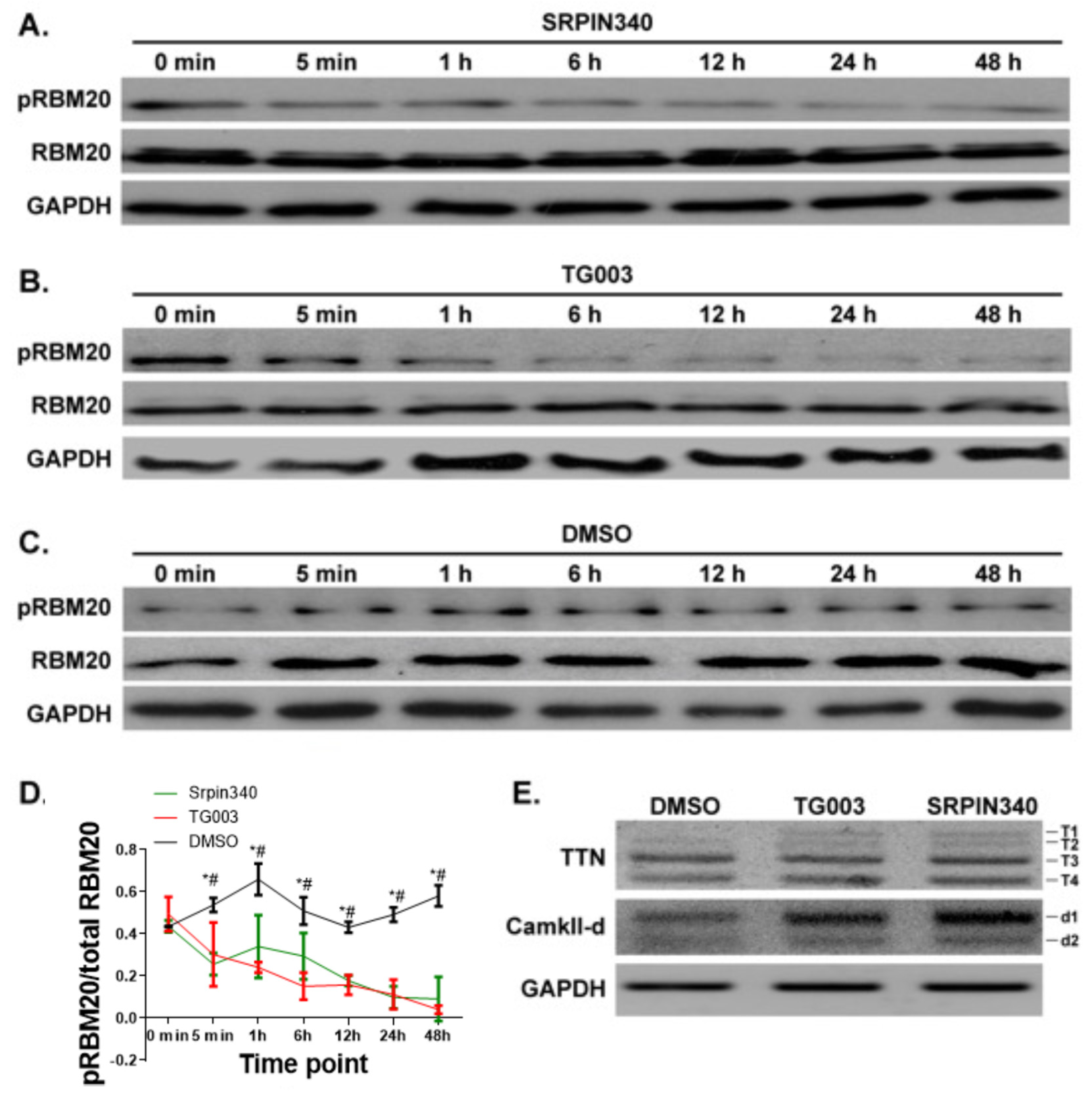
3.4. Inhibition of CLK and SRPK Family Kinases Reduces RBM20 RSRSP Phosphorylation and Leads to Aberrant Pre-mRNA Splicing in NRCMs
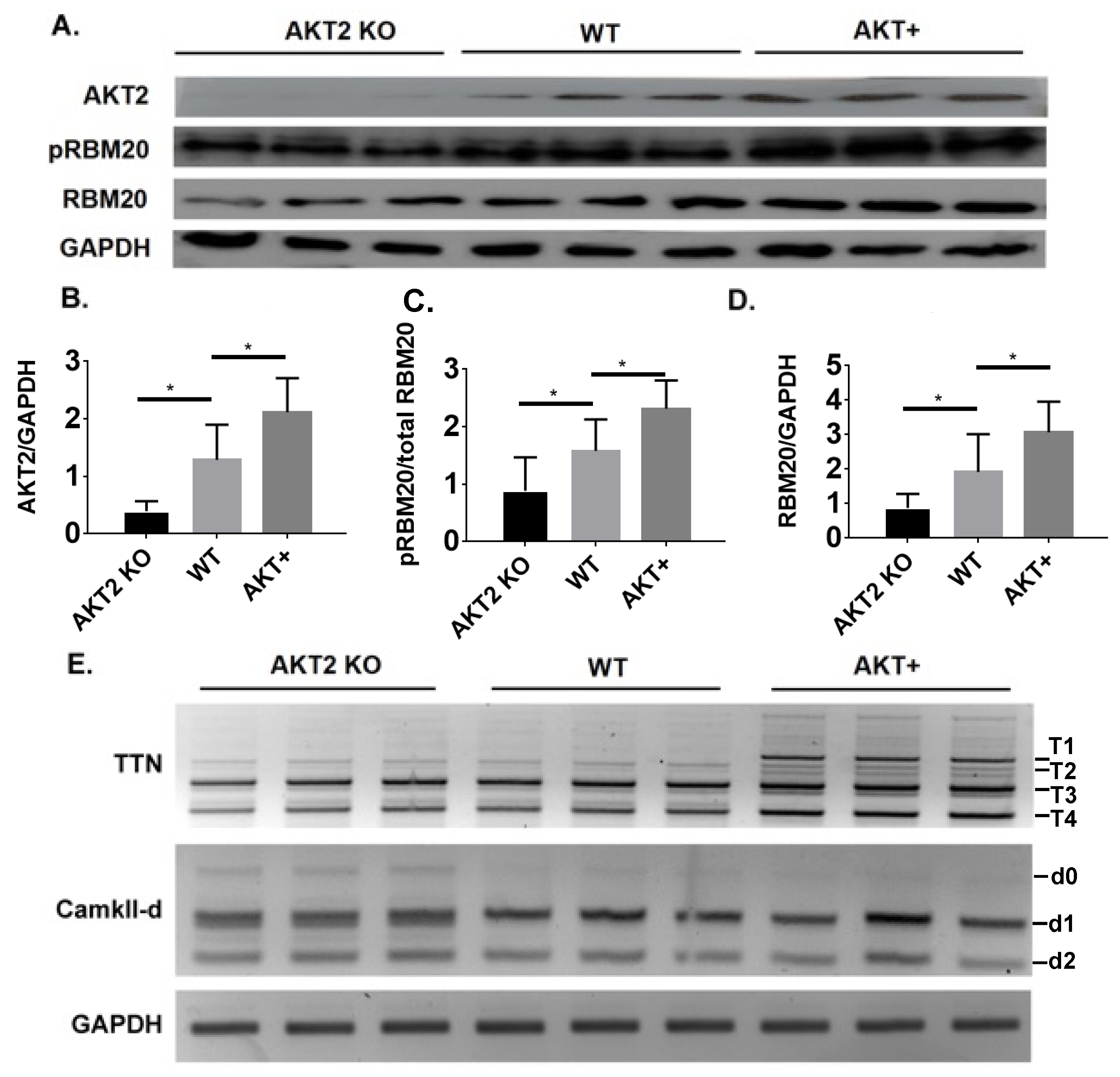
3.5. Phosphorylation of the RSRSP Stretch in RBM20 and Pre-mRNA Splicing of RBM20 Targets Are Altered in Akt2 KO and Transgenic Mice
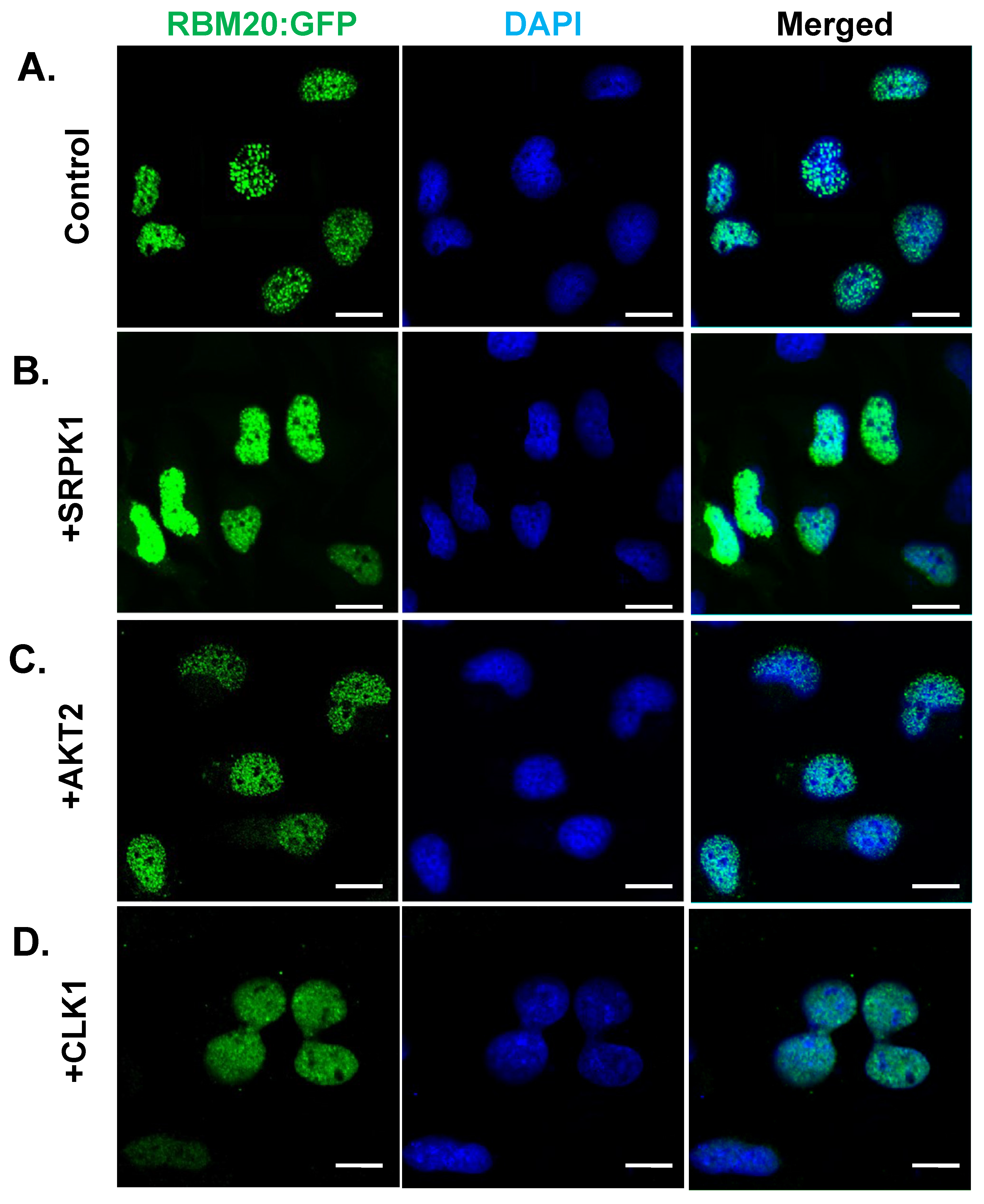
3.6. Overexpression of CLK1, SRPK1, or AKT2 Alone Does Not Facilitate RBM20 Nucleocytoplasmic Transport
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greaser, M.L.; Warren, C.M.; Esbona, K.; Guo, W.; Duan, Y.; Parrish, A.M.; Krzesinski, P.R.; Norman, H.S.; Dunning, S.; Fitzsimons, D.P.; et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J. Mol. Cell. Cardiol. 2008, 44, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Pleitner, J.M.; Saupe, K.W.; Greaser, M.L. Pathophysiological defects and transcriptional profiling in the RBM20-/- rat model. PLoS ONE 2013, 8, e84281. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Schafer, S.; Greaser, M.L.; Radke, M.H.; Liss, M.; Govindarajan, T.; Maatz, H.; Schulz, H.; Li, S.; Parrish, A.M.; et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 2012, 18, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Greaser, M.L.; Krzesinski, P.R.; Warren, C.M.; Kirkpatrick, B.; Campbell, K.S.; Moss, R.L. Developmental changes in rat cardiac titin/connectin: Transitions in normal animals and in mutants with a delayed pattern of isoform transition. J. Muscle Res. Cell Motil. 2005, 26, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Guo, W.; Dewey, C.N.; Greaser, M.L. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013, 41, 2659–2672. [Google Scholar] [CrossRef]
- Freiburg, A.; Gautel, M. A Molecular Map of the Interactions between Titin and Myosin-Binding Protein C: Implications for Sarcomeric Assembly in Familial Hypertrophic Cardiomyopathy. Eur. J. Biochem. 1996, 235, 317–323. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004, 94, 284–295. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. Titin and its associated proteins: The third myofilament system of the sarcomere. Adv. Protein Chem. 2005, 71, 89–119. [Google Scholar] [CrossRef]
- Granzier, H.L.; Labeit, S. The giant muscle protein titin is an adjustable molecular spring. Exerc. Sport Sci. Rev. 2006, 34, 50–53. [Google Scholar] [CrossRef]
- Krüger, M.; Linke, W.A. The giant protein titin: A regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 2011, 286, 9905–9912. [Google Scholar] [CrossRef] [Green Version]
- Linke, W.A. Sense and stretchability: The role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res. 2008, 77, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Sun, M. RBM20, a potential target for treatment of cardiomyopathy via titin isoform switching. Biophys. Rev. 2018, 10, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Maatz, H.; Jens, M.; Liss, M.; Schafer, S.; Heinig, M.; Kirchner, M.; Adami, E.; Rintisch, C.; Dauksaite, V.; Radke, M.H.; et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J. Clin. Investig. 2014, 124, 3419–3430. [Google Scholar] [CrossRef]
- van den Hoogenhof, M.M.G.; Beqqali, A.; Amin, A.S.; van der Made, I.; Aufiero, S.; Khan, M.A.F.; Schumacher, C.A.; Jansweijer, J.A.; van Spaendonck-Zwarts, K.Y.; Remme, C.A.; et al. RBM20 Mutations Induce an Arrhythmogenic Dilated Cardiomyopathy Related to Disturbed Calcium Handling. Circulation 2018, 138, 1330–1342. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhu, C.; Yin, Z.; Wang, Q.; Sun, M.; Cao, H.; Greaser, M.L. Splicing Factor RBM20 Regulates Transcriptional Network of Titin Associated and Calcium Handling Genes in The Heart. Int. J. Biol. Sci. 2018, 14, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Methawasin, M.; Hutchinson, K.R.; Lee, E.-J.; Smith, J.E., III; Saripalli, C.; Hidalgo, C.G.; Ottenheijm, C.A.; Granzier, H.J.C. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation 2014, 129, 1924–1936. [Google Scholar] [CrossRef] [PubMed]
- Brauch, K.M.; Karst, M.L.; Herron, K.J.; de Andrade, M.; Pellikka, P.A.; Rodeheffer, R.J.; Michels, V.V.; Olson, T.M. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol. 2009, 54, 930–941. [Google Scholar] [CrossRef]
- Millat, G.; Bouvagnet, P.; Chevalier, P.; Sebbag, L.; Dulac, A.; Dauphin, C.; Jouk, P.S.; Delrue, M.A.; Thambo, J.B.; Le Metayer, P.; et al. Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur. J. Med. Genet. 2011, 54, e570–e575. [Google Scholar] [CrossRef]
- Monaco, I.; Santacroce, R.; Casavecchia, G.; Correale, M.; Bottigliero, D.; Cordisco, G.; Leccese, A.; Di Biase, M.; Margaglione, M.; Brunetti, N.D. Double de novo mutations in dilated cardiomyopathy with cardiac arrest. J. Electrocardiol. 2019, 53, 40–43. [Google Scholar] [CrossRef]
- Pantou, M.P.; Gourzi, P.; Gkouziouta, A.; Tsiapras, D.; Zygouri, C.; Constantoulakis, P.; Adamopoulos, S.; Degiannis, D.J.C. Phenotypic heterogeneity within members of a family carrying the same RBM20 mutation R634W. Cardiology 2018, 141, 150–155. [Google Scholar] [CrossRef]
- Refaat, M.M.; Lubitz, S.A.; Makino, S.; Islam, Z.; Frangiskakis, J.M.; Mehdi, H.; Gutmann, R.; Zhang, M.L.; Bloom, H.L.; MacRae, C.A. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm 2012, 9, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Rampersaud, E.; Siegfried, J.D.; Norton, N.; Li, D.; Martin, E.; Hershberger, R.E. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog. Pediatr. Cardiol. 2011, 31, 39–47. [Google Scholar] [CrossRef]
- Li, D.; Morales, A.; Gonzalez-Quintana, J.; Norton, N.; Siegfried, J.D.; Hofmeyer, M.; Hershberger, R.E. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci. 2010, 3, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.W.; Oommen, S.; Qureshi, M.Y.; Goetsch, S.C.; Pease, D.R.; Sundsbak, R.S.; Guo, W.; Sun, M.; Sun, H.; Kuroyanagi, H.; et al. Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat. Med. 2020, 26, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Ihara, K.; Sasano, T.; Hiraoka, Y.; Togo-Ohno, M.; Soejima, Y.; Sawabe, M.; Tsuchiya, M.; Ogawa, H.; Furukawa, T.; Kuroyanagi, H. A missense mutation in the RSRSP stretch of Rbm20 causes dilated cardiomyopathy and atrial fibrillation in mice. Sci. Rep. 2020, 10, 17894. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Sun, M.; Jin, Y.; Braz, C.U.; Khatib, H.; Hacker, T.A.; Liss, M.; Gotthardt, M.; Granzier, H.; et al. RBM20 phosphorylation and its role in nucleocytoplasmic transport and cardiac pathogenesis. FASEB J. 2022, 36, e22302. [Google Scholar] [CrossRef]
- Serrano, P.; Aubol, B.E.; Keshwani, M.M.; Forli, S.; Ma, C.-T.; Dutta, S.K.; Geralt, M.; Wüthrich, K.; Adams, J.A. Directional phosphorylation and nuclear transport of the splicing factor SRSF1 is regulated by an RNA recognition motif. J. Mol. Biol. 2016, 428, 2430–2445. [Google Scholar] [CrossRef]
- Zhu, C.; Chen, Z.; Guo, W. Pre-mRNA mis-splicing of sarcomeric genes in heart failure. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2056–2063. [Google Scholar] [CrossRef]
- Rexiati, M.; Sun, M.; Guo, W. Muscle-Specific Mis-Splicing and Heart Disease Exemplified by RBM20. Genes 2018, 9, 18. [Google Scholar] [CrossRef]
- Ghosh, G.; Adams, J.A. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011, 278, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Jin, Y.; Zhu, C.; Zhang, Y.; Liss, M.; Gotthardt, M.; Ren, J.; Ge, Y.; Guo, W. RBM20 phosphorylation on serine/arginine domain is crucial to regulate pre-mRNA splicing and protein shuttling in the heart. bioRxiv 2020, 297002. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Sette, C. Phosphorylation-mediated regulation of alternative splicing in cancer. Int. J. Cell Biol. 2013, 2013, 151839. [Google Scholar] [CrossRef] [PubMed]
- Prasad, J.; Colwill, K.; Pawson, T.; Manley, J.L. The protein kinase Clk/Sty directly modulates SR protein activity: Both hyper-and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999, 19, 6991–7000. [Google Scholar] [CrossRef] [PubMed]
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 242. [Google Scholar] [CrossRef]
- Lai, M.C.; Lin, R.I.; Tarn, W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 10154–10159. [Google Scholar] [CrossRef]
- Mermoud, J.E.; Cohen, P.T.; Lamond, A.I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994, 13, 5679–5688. [Google Scholar] [CrossRef]
- Huang, Y.; Yario, T.A.; Steitz, J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 2004, 101, 9666–9670. [Google Scholar] [CrossRef]
- Lai, M.-C.; Tarn, W.-Y. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 2004, 279, 31745–31749. [Google Scholar] [CrossRef]
- Murayama, R.; Kimura-Asami, M.; Togo-Ohno, M.; Yamasaki-Kato, Y.; Naruse, T.K.; Yamamoto, T.; Hayashi, T.; Ai, T.; Spoonamore, K.G.; Kovacs, R.J.; et al. Phosphorylation of the RSRSP stretch is critical for splicing regulation by RNA-Binding Motif Protein 20 (RBM20) through nuclear localization. Sci. Rep. 2018, 8, 8970. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Yang, L.; Zhu, L.; Xu, X.; Ceylan, A.F.; Guo, W.; Yang, J.; Zhang, Y. Akt2 ablation prolongs life span and improves myocardial contractile function with adaptive cardiac remodeling: Role of Sirt1-mediated autophagy regulation. Aging Cell 2017, 16, 976–987. [Google Scholar] [CrossRef]
- Zhu, C.; Guo, W. Detection and quantification of the giant protein titin by SDS-agarose gel electrophoresis. MethodsX 2017, 4, 320–327. [Google Scholar] [CrossRef]
- Zhu, C.; Yin, Z.; Ren, J.; McCormick, R.J.; Ford, S.P.; Guo, W. RBM20 is an essential factor for thyroid hormone-regulated titin isoform transition. J. Mol. Cell Biol. 2015, 7, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yin, Z.; Tan, B.; Guo, W. Insulin regulates titin pre-mRNA splicing through the PI3K-Akt-mTOR kinase axis in a RBM20-dependent manner. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Anastassiadis, T.; Deacon, S.W.; Devarajan, K.; Ma, H.; Peterson, J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Zhang, J.; de Lange, W.J.; Gregorich, Z.R.; Karp, H.; Farrell, E.T.; Mitchell, S.D.; Tucholski, T.; Lin, Z.; Biermann, M. An unbiased proteomics method to assess the maturation of human pluripotent stem cell–derived cardiomyocytes. Circ. Res. 2019, 125, 936–953. [Google Scholar] [CrossRef]
- Lin, Z.; Wei, L.; Cai, W.; Zhu, Y.; Tucholski, T.; Mitchell, S.D.; Guo, W.; Ford, S.P.; Diffee, G.M.; Ge, Y. Simultaneous Quantification of Protein Expression and Modifications by Top-down Targeted Proteomics: A Case of the Sarcomeric Subproteome. Mol. Cell. Proteom. 2019, 18, 594–605. [Google Scholar] [CrossRef]
- Jin, Y.; Lin, Z.; Xu, Q.; Fu, C.; Zhang, Z.; Zhang, Q.; Pritts, W.A.; Ge, Y. Comprehensive characterization of monoclonal antibody by Fourier transform ion cyclotron resonance mass spectrometry. MAbs 2019, 11, 106–115. [Google Scholar] [CrossRef]
- Liu, X.; Hengel, S.; Wu, S.; Tolic, N.; Pasa-Tolic, L.; Pevzner, P.A. Identification of ultramodified proteins using top-down tandem mass spectra. J. Proteome Res. 2013, 12, 5830–5838. [Google Scholar] [CrossRef]
- Becich, M.J. Advancing Practice, Instruction, and Innovation Through Informatics (APIII 2006): Scientific Session Presentation Abstracts and Scientific Poster Session Abstracts. Arch. Pathol. Lab. Med. 2007, 131, 805. [Google Scholar]
- Cai, W.; Guner, H.; Gregorich, Z.R.; Chen, A.J.; Ayaz-Guner, S.; Peng, Y.; Valeja, S.G.; Liu, X.; Ge, Y. MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics. Mol. Cell. Proteom. 2016, 15, 703–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Sun, M.; Zhou, H.; Ajoolabady, A.; Zhou, Y.; Tao, J.; Sowers, J.R.; Zhang, Y. FUNDC1 interacts with FBXL2 to govern mitochondrial integrity and cardiac function through an IP3R3-dependent manner in obesity. Sci. Adv. 2020, 6, eabc8561. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, J.F.; Screaton, G.R.; Krainer, A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998, 12, 55–66. [Google Scholar] [CrossRef]
- Hertel, K.J.; Graveley, B.R. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 2005, 30, 115–118. [Google Scholar] [CrossRef]
- Wu, J.Y.; Maniatis, T.J.C. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 1993, 75, 1061–1070. [Google Scholar] [CrossRef]
- Lin, S.; Fu, X.D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007, 623, 107–122. [Google Scholar] [CrossRef]
- Liu, Q.; Dreyfuss, G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995, 15, 2800–2808. [Google Scholar] [CrossRef]
- Yun, C.Y.; Fu, X.-D. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 2000, 150, 707–718. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Edmond, V.; Moysan, E.; Khochbin, S.; Matthias, P.; Brambilla, C.; Brambilla, E.; Gazzeri, S.; Eymin, B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011, 30, 510–523. [Google Scholar] [CrossRef]
- Kannan, N.; Neuwald, A.F. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2α. Protein Sci. 2004, 13, 2059–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Patel, N.A.; Watson, J.E.; Apostolatos, H.; Kleiman, E.; Hanson, O.; Hagiwara, M.; Cooper, D.R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβII messenger ribonucleic acid. Endocrinology 2009, 150, 2087–2097. [Google Scholar] [CrossRef]
- Blaustein, M.; Pelisch, F.; Tanos, T.; Munoz, M.J.; Wengier, D.; Quadrana, L.; Sanford, J.R.; Muschietti, J.P.; Kornblihtt, A.R.; Caceres, J.F.; et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005, 12, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.A.; Kaneko, S.; Apostolatos, H.S.; Bae, S.S.; Watson, J.E.; Davidowitz, K.; Chappell, D.S.; Birnbaum, M.J.; Cheng, J.Q.; Cooper, D.R. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J. Biol. Chem. 2005, 280, 14302–14309. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T.; Hosoya, T.; Shimizu, S.; Sumi, K.; Oshiro, T.; Yoshinaka, Y.; Suzuki, M.; Yamamoto, N.; Herzenberg, L.A.; Herzenberg, L.A.; et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA 2006, 103, 11329–11333. [Google Scholar] [CrossRef]
- Hayes, G.M.; Carrigan, P.E.; Beck, A.M.; Miller, L.J. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006, 66, 3819–3827. [Google Scholar] [CrossRef]
- Yomoda, J.; Muraki, M.; Kataoka, N.; Hosoya, T.; Suzuki, M.; Hagiwara, M.; Kimura, H. Combination of Clk family kinase and SRp75 modulates alternative splicing of Adenovirus E1A. Genes Cells 2008, 13, 233–244. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Ding, J.H.; Adams, J.A.; Ghosh, G.; Fu, X.D. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009, 23, 482–495. [Google Scholar] [CrossRef]
- Bull, M.; Methawasin, M.; Strom, J.; Nair, P.; Hutchinson, K.; Granzier, H. Alternative Splicing of Titin Restores Diastolic Function in an HFpEF-Like Genetic Murine Model (TtnDeltaIAjxn). Circ. Res. 2016, 119, 764–772. [Google Scholar] [CrossRef]
- Mermoud, J.E.; Cohen, P.; Lamond, A.I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992, 20, 5263–5269. [Google Scholar] [CrossRef]
- Stojdl, D.F.; Bell, J.C. SR protein kinases: The splice of life. Biochem. Cell Biol. 1999, 77, 293–298. [Google Scholar] [CrossRef]
- Xiao, S.-H.; Manley, J.L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997, 11, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Hoang, A.; Sinha, R.; Zhong, X.-Y.; Fu, X.-D.; Krainer, A.R.; Ghosh, G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 8233–8238. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Feng, Y.; Manley, J.L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 2004, 427, 553–558. [Google Scholar] [CrossRef]
- Shin, C.; Manley, J.L. The SR protein SRp38 represses splicing in M phase cells. Cell 2002, 111, 407–417. [Google Scholar] [CrossRef]
- Lai, M.C.; Lin, R.I.; Huang, S.Y.; Tsai, C.W.; Tarn, W.Y. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2000, 275, 7950–7957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.; Jin, Y.; Zhang, Y.; Gregorich, Z.R.; Ren, J.; Ge, Y.; Guo, W. SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20. Genes 2022, 13, 1526. https://doi.org/10.3390/genes13091526
Sun M, Jin Y, Zhang Y, Gregorich ZR, Ren J, Ge Y, Guo W. SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20. Genes. 2022; 13(9):1526. https://doi.org/10.3390/genes13091526
Chicago/Turabian StyleSun, Mingming, Yutong Jin, Yanghai Zhang, Zachery R Gregorich, Jun Ren, Ying Ge, and Wei Guo. 2022. "SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20" Genes 13, no. 9: 1526. https://doi.org/10.3390/genes13091526
APA StyleSun, M., Jin, Y., Zhang, Y., Gregorich, Z. R., Ren, J., Ge, Y., & Guo, W. (2022). SR Protein Kinases Regulate the Splicing of Cardiomyopathy-Relevant Genes via Phosphorylation of the RSRSP Stretch in RBM20. Genes, 13(9), 1526. https://doi.org/10.3390/genes13091526





