Alterations in Microbiota and Metabolites Related to Spontaneous Diabetes and Pre-Diabetes in Rhesus Macaques
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. 16S rRNA Gene Sequencing Analysis
2.3. Metabolomics Analysis
2.4. Correlation Analysis
3. Result
3.1. Clinical Characteristics
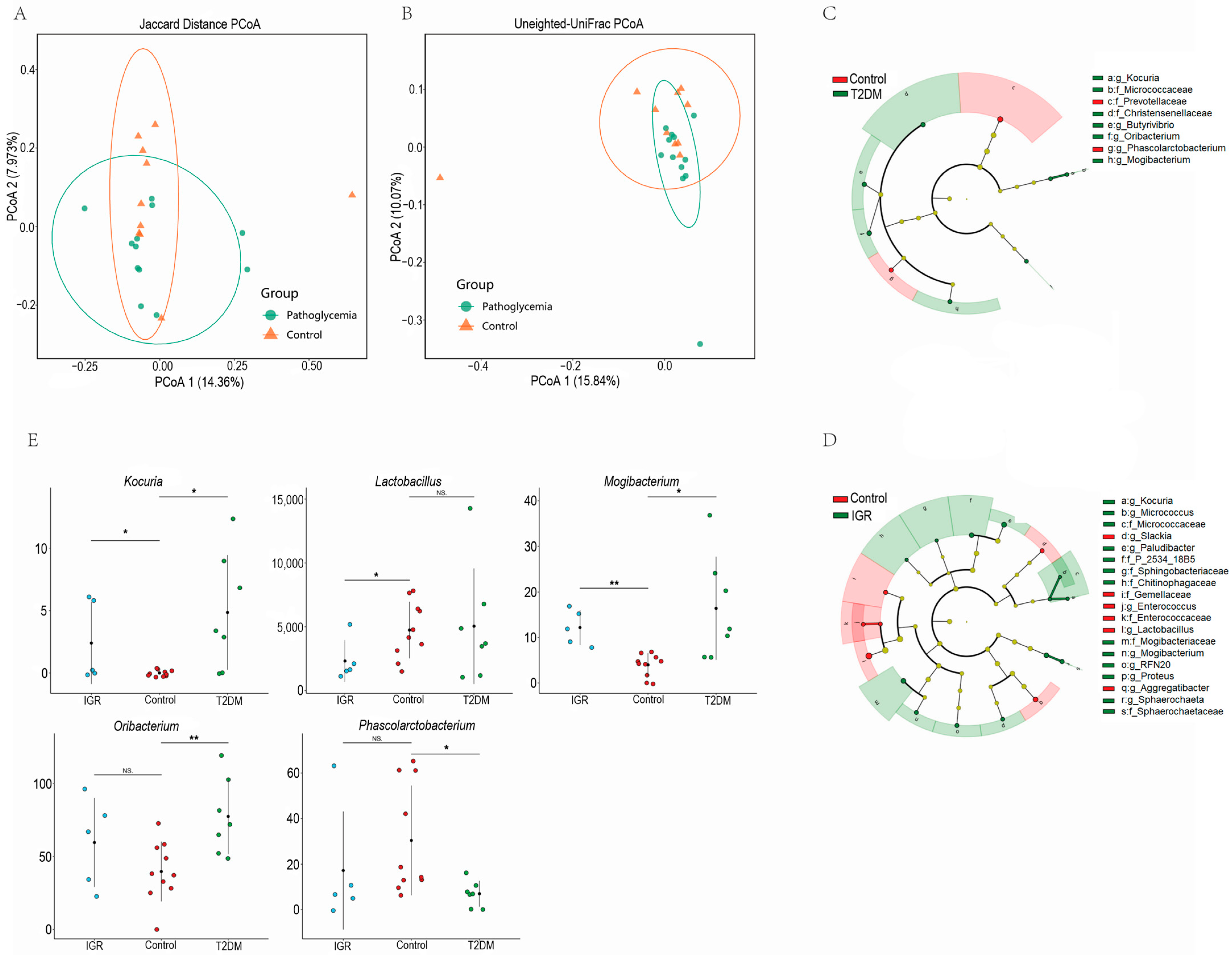
3.2. Differences of Gut Microbial Composition between T2DM, IGR Macaques and Healthy Subjects
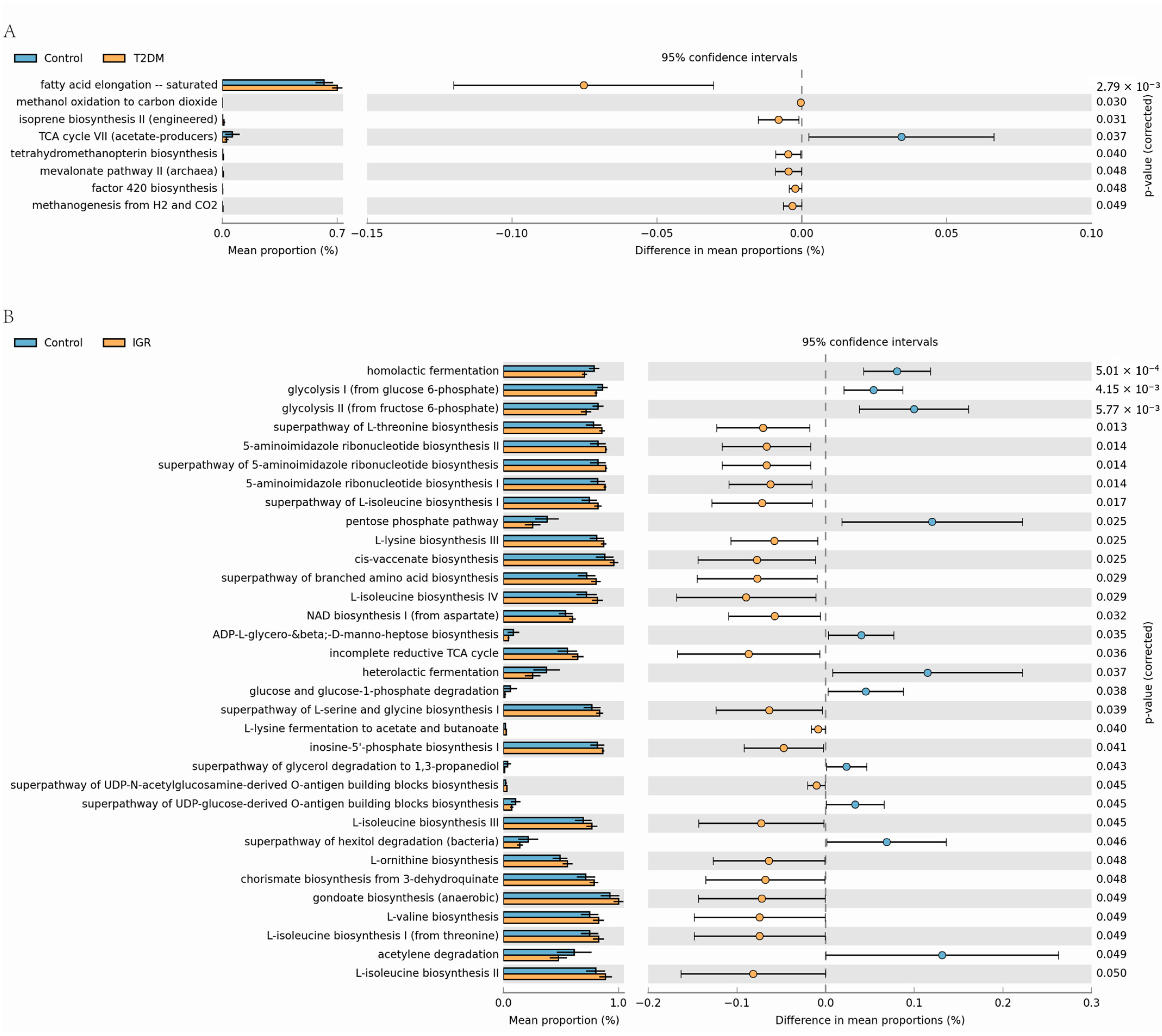
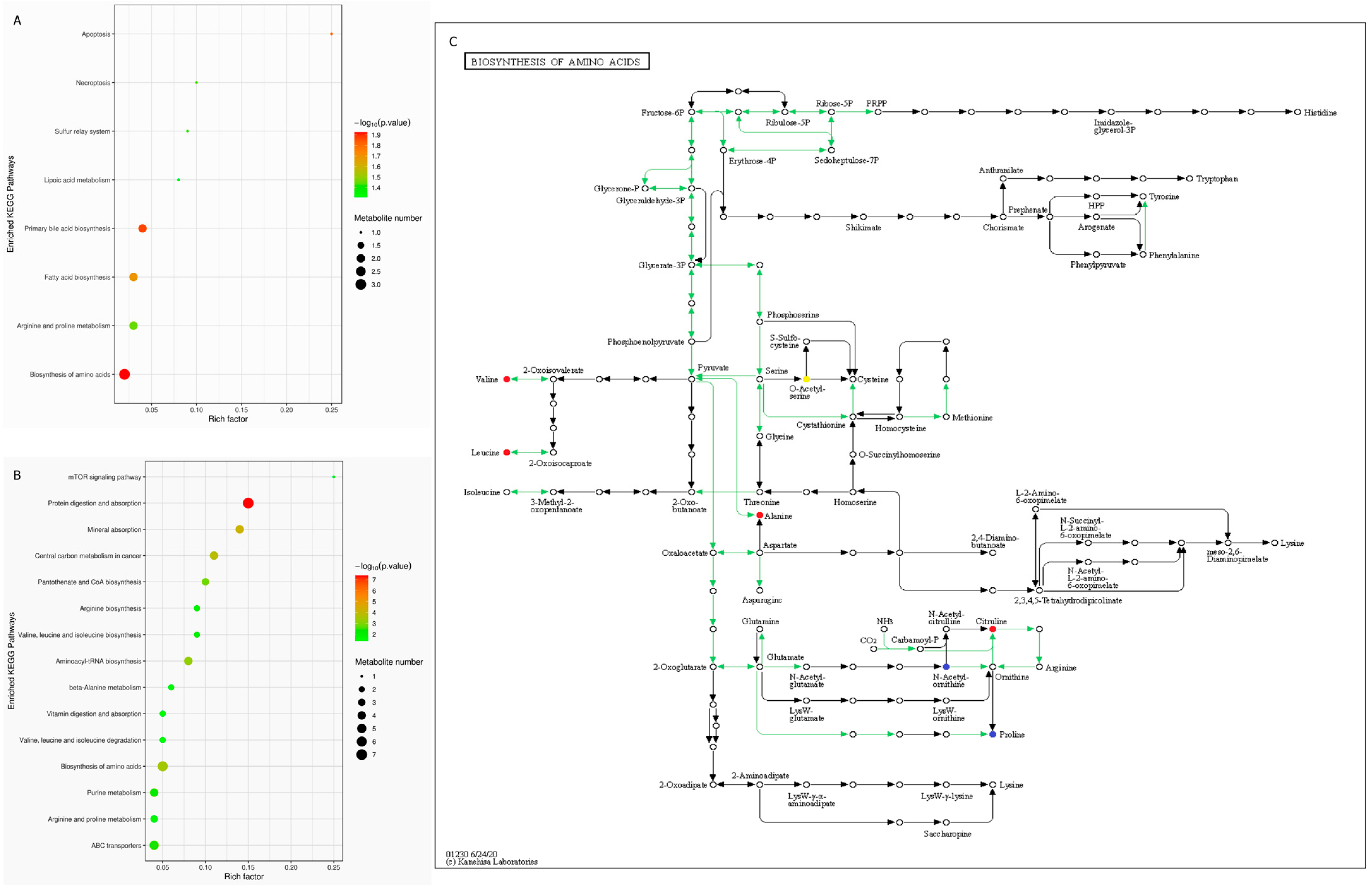
3.3. Fecal Metabolite Profiles in T2DM, IGR Macaques and Healthy Subjects
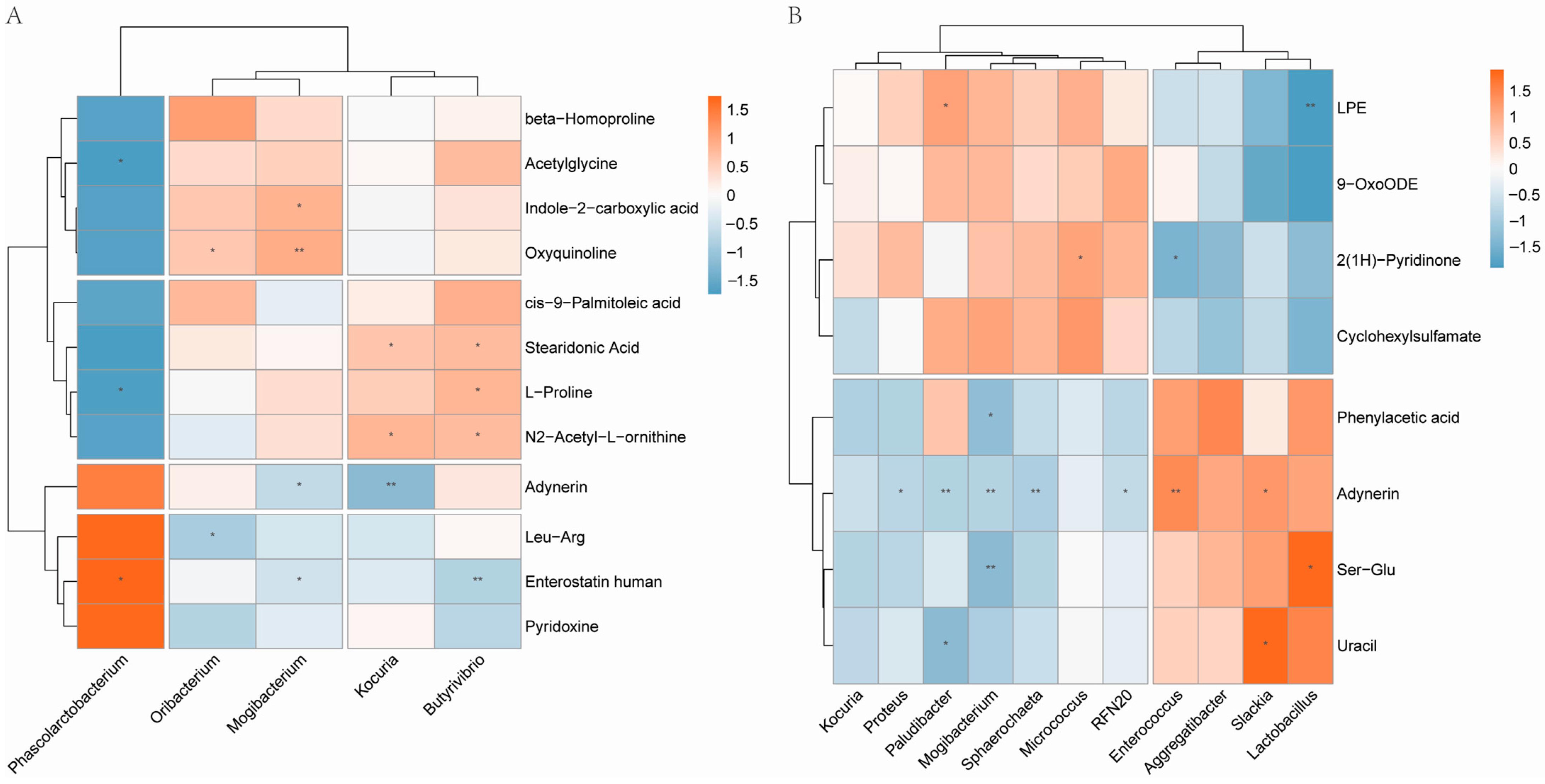
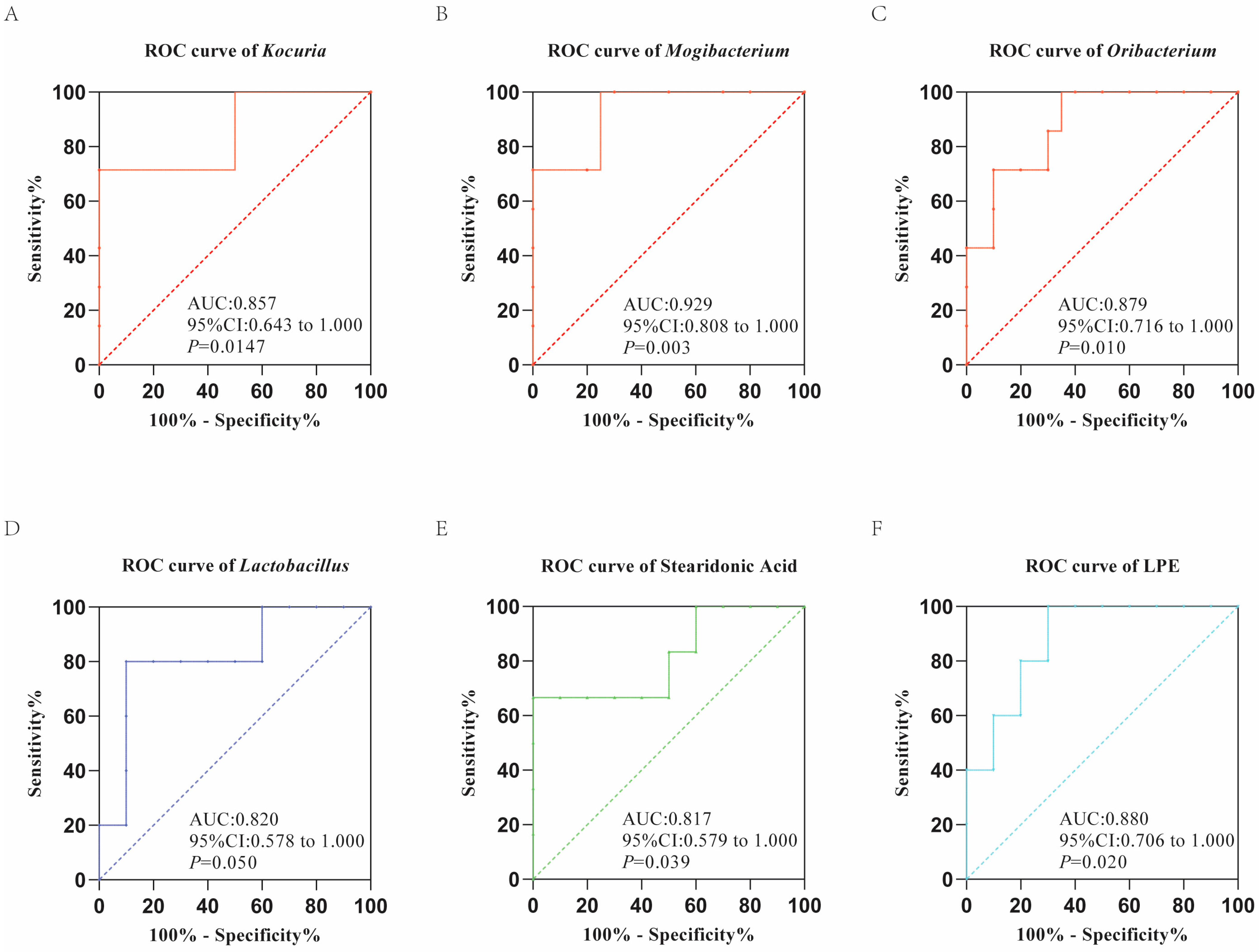
3.4. Correlation Analysis of Gut Microbiota and Fecal Metabolic Traits
4. Discussion
4.1. Hyperglycemia and High Insulin Resistance in T2DM and IGR Macaques
4.2. Glucose Metabolism Related Bacteria Differed in T2DM and IGR Macaques
4.3. Production of Bile Acids Reduced in T2DM Macaques
4.4. A Variety of Fatty Acids Differed in T2DM and IGR Macaques
4.5. BCAAs Metabolism Enhanced in T2DM and IGR Macaques
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Burke, J.; Kovacs, B.; Barton, L.; Sander, S. Health Care Utilization and Costs in Type 2 Diabetes Mellitus and Their Association with Renal Impairment. Postgrad. Med. 2012, 124, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Federation, I.D. IDF Diabetes Atlas, 9th ed. Available online: https://www.diabetesatlas.org (accessed on 1 August 2020).
- American Diabetes, A. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2021. Diabetes Care 2021, 44, S15–S33. [Google Scholar] [CrossRef] [PubMed]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770–775. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Interactions Between Gut Microbiota and Host Metabolism Predisposing to Obesity and Diabetes. Annu. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Abu Al-Soud, W.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Yassour, M.; Lim, M.Y.; Yun, H.S.; Tickle, T.L.; Sung, J.; Song, Y.M.; Lee, K.; Franzosa, E.A.; Morgan, X.C.; Gevers, D.; et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016, 8, 17. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Zhang, J.T.; Cai, Y.J.; Cheng, T.L.; Cheng, C.; Wang, Y.; Zhang, C.C.; Nie, Y.H.; Chen, Z.F.; et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 2016, 530, 98–102. [Google Scholar] [CrossRef]
- Antony, J.M.; MacDonald, K.S. A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine 2015, 33, 3073–3083. [Google Scholar] [CrossRef]
- Davey, R.T.; Dodd, L.; Proschan, M.A.; Neaton, J.; Nordwall, J.N.; Koopmeiners, J.S.; Beigel, J.; Tierney, J.; Lane, H.C.; Fauci, A.S.; et al. A Randomized, Controlled Trial of ZMapp for Ebola Virus Infection. N. Engl. J. Med. 2016, 375, 1448–1456. [Google Scholar] [CrossRef]
- Wang, J.L.; Xu, S.M.; Gao, J.; Zhang, L.Q.; Zhang, Z.G.; Yang, W.H.; Li, Y.H.; Liao, S.S.; Zhou, H.; Liu, P.S.; et al. SILAC-based quantitative proteomic analysis of the livers of spontaneous obese and diabetic rhesus monkeys. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E294–E306. [Google Scholar] [CrossRef]
- McCurdy, C.E.; Bishop, J.M.; Williams, S.M.; Grayson, B.E.; Smith, M.S.; Friedman, J.E.; Grove, K.L. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J. Clin. Investig. 2009, 119, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Gong, L.; Yang, Z.Y.; Chen, W.; Chen, Y.S.; Xu, Z.Q.; Wu, B.; Tang, C.G.; Gao, F.B.; Zeng, W. Diastolic dysfunction in spontaneous type 2 diabetes rhesus monkeys: A study using echocardiography and magnetic resonance imaging. BMC Cardiovasc. Disord. 2015, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Cefalu, W.T. Animal models of type 2 diabetes: Clinical presentation and pathophysiological relevance to the human condition. ILAR J. 2006, 47, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Kleinert, M.; Clemmensen, C.; Hofmann, S.M.; Moore, M.C.; Renner, S.; Woods, S.C.; Huypens, P.; Beckers, J.; de Angelis, M.H.; Schurmann, A.; et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 2018, 14, 140–162. [Google Scholar] [CrossRef]
- Gong, L.; Zeng, W.; Yang, Z.Y.; Chen, Z.L.; Cheng, A.C.; Shen, Y.B.; Zeng, L.C.; Luo, Q.H.; Yang, Y. Comparison of the Clinical Manifestations of Type 2 Diabetes Mellitus Between Rhesus Monkey (Macaca mulatta lasiotis) and Human Being. Pancreas 2013, 42, 537–542. [Google Scholar] [CrossRef]
- Taborsky, G.J.; Ahren, B.; Havel, P.J. Autonomic mediation of glucagon secretion during hypoglycemia—Implications for impaired α-cell responses in type 1 diabetes. Diabetes 1998, 47, 995–1005. [Google Scholar] [CrossRef]
- Ahren, B.; Baldwin, R.M.; Havel, P.J. Pharmacokinetics of human leptin in mice and rhesus monkeys. Int. J. Obes. 2000, 24, 1579–1585. [Google Scholar] [CrossRef]
- D’Alessio, D.A.; Kieffer, T.J.; Taborsky, G.J.; Havel, P.J. Activation of the parasympathetic nervous system is necessary for normal meal-induced insulin secretion in rhesus macaques. J. Clin. Endocrinol. Metab. 2001, 86, 1253–1259. [Google Scholar] [CrossRef]
- Adams, S.H.; Stanhope, K.L.; Grant, R.W.; Cummings, B.P.; Havel, P.J. Metabolic and endocrine profiles in response to systemic infusion of fructose and glucose in rhesus macaques. Endocrinology 2008, 149, 3002–3008. [Google Scholar] [CrossRef][Green Version]
- Swarbrick, M.M.; Havel, P.J.; Levin, A.A.; Bremer, A.A.; Stanhope, K.L.; Butler, M.; Booten, S.L.; Graham, J.L.; Mckay, R.A.; Murray, S.F.; et al. Inhibition of Protein Tyrosine Phosphatase-1B with Antisense Oligonucleotides Improves Insulin Sensitivity and Increases Adiponectin Concentrations in Monkeys. Endocrinology 2009, 150, 1670–1679. [Google Scholar] [CrossRef]
- Pound, L.D.; Kievit, P.; Grove, K.L. The nonhuman primate as a model for type 2 diabetes. Curr. Opin. Endocrinol. 2014, 21, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Bremer, A.A.; Stanhope, K.L.; Graham, J.L.; Cummings, B.P.; Wang, W.L.; Saville, B.R.; Havel, P.J. Fructose-Fed Rhesus Monkeys: A Nonhuman Primate Model of Insulin Resistance, Metabolic Syndrome, and Type 2 Diabetes. Clin. Transl. Sci. 2011, 4, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Cowie, C.C.; Rust, K.F.; Byrd-Holt, D.D.; Gregg, E.W.; Ford, E.S.; Geiss, L.S.; Bainbridge, K.E.; Fradkin, J.E. Prevalence of Diabetes and High Risk for Diabetes Using A1C Criteria in the US Population in 1988–2006. Diabetes Care 2010, 33, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.; Hazuda, H.P.; Haffner, S.M. Insulin resistance and excess risk of diabetes in Mexican-Americans: The San Antonio Heart Study. J. Clin. Endocrinol. Metab. 2012, 97, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Minh, C.C.; Chung, J.; Kozyrakis, C.; Olukotun, K. STAMP: Stanford Transactional Applications for Multi-Processing. In 2008 IEEE International Symposium on Workload Characterization; IEEE: Piscataway, NJ, USA, 2008; pp. 31–42. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.Z.; Bourque, G.; Wishart, D.S.; Xia, J.G. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Mitteer, D.R.; Greer, B.D.; Randall, K.R.; Briggs, A.M. Further Evaluation of Teaching Behavior Technicians to Input Data and Graph Using GraphPad Prism. Behav. Anal. 2020, 20, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Prentki, M.; Joly, E.; El-Assaad, W.; Roduit, R. Malonyl-CoA signaling, lipid partitioning, and glucolipotoxicity: Role in β-cell adaptation and failure in the etiology of diabetes. Diabetes 2002, 51, S405–S413. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.P. Chronic oxidative stress as a central mechanism for glucose toxicity in pancreatic islet β cells in diabetes. J. Biol. Chem. 2004, 279, 42351–42354. [Google Scholar] [CrossRef]
- Ahlqvist, E.; Storm, P.; Karajamaki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef]
- Sladek, R. The many faces of diabetes: Addressing heterogeneity of a complex disease. Lancet Diabetes Endocrinol. 2018, 6, 348–349. [Google Scholar] [CrossRef]
- Locke, A.E.; Steinberg, K.M.; Chiang, C.W.K.; Service, S.K.; Havulinna, A.S.; Stell, L.; Pirinen, M.; Abel, H.J.; Chiang, C.C.; Fulton, R.S.; et al. Author Correction: Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 2019, 575, E4. [Google Scholar] [CrossRef]
- Schnurr, T.M.; Jakupovic, H.; Carrasquilla, G.D.; Angquist, L.; Grarup, N.; Sorensen, T.I.A.; Tjonneland, A.; Overvad, K.; Pedersen, O.; Hansen, T.; et al. Obesity, unfavourable lifestyle and genetic risk of type 2 diabetes: A case-cohort study. Diabetologia 2020, 63, 1324–1332. [Google Scholar] [CrossRef]
- Tritt, M.; Sgouroudis, E.; d’Hennezel, E.; Albanese, A.; Piccirillo, C.A. Functional waning of naturally occurring CD4+ regulatory T-cells contributes to the onset of autoimmune diabetes. Diabetes 2008, 57, 113–123. [Google Scholar] [CrossRef]
- Bapat, S.P.; Myoung Suh, J.; Fang, S.; Liu, S.; Zhang, Y.; Cheng, A.; Zhou, C.; Liang, Y.; LeBlanc, M.; Liddle, C.; et al. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature 2015, 528, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E.C.; Walter, J.; Backhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Sanna, S.; van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vila, A.V.; Vosa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.M.A.E.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef]
- Robertson, R.P.; Zhou, H.R.; Zhang, T.; Harmon, J.S. Chronic oxidative stress as a mechanism for glucose toxicity of the β cell in Type 2 diabetes. Cell Biochem. Biophys. 2007, 48, 139–146. [Google Scholar] [CrossRef]
- Lowell, B.B.; Shulmanz, G.I. Mitochond rial dysfunction and type 2 diabetes. Science 2005, 307, 384–387. [Google Scholar] [CrossRef]
- Ma, Z.A.; Zhao, Z.S.; Turk, J. Mitochondrial Dysfunction and β-Cell Failure in Type 2 Diabetes Mellitus. Exp. Diabetes Res. 2012, 2012, 703538. [Google Scholar] [CrossRef]
- Angelakis, E.; Armougom, F.; Carriere, F.; Bachar, D.; Laugier, R.; Lagier, J.C.; Robert, C.; Michelle, C.; Henrissat, B.; Raoult, D. A Metagenomic Investigation of the Duodenal Microbiota Reveals Links with Obesity. PLoS ONE 2015, 10, e0137784. [Google Scholar] [CrossRef]
- Petrov, P.D.; Garcia-Mediavilla, M.V.; Guzman, C.; Porras, D.; Nistal, E.; Martinez-Florez, S.; Castell, J.V.; Gonzalez-Gallego, J.; Sanchez-Campos, S.; Jover, R. A Network Involving Gut Microbiota, Circulating Bile Acids, and Hepatic Metabolism Genes That Protects Against Non-Alcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2019, 63, e1900487. [Google Scholar] [CrossRef]
- Vallim, T.Q.D.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Zhao, L.J.; Lou, H.X.; Peng, Y.; Chen, S.H.; Zhang, Y.L.; Li, X.B. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Martins, R.; Sullivan, M.C.; Friedman, E.S.; Misic, A.M.; El-Fahmawi, A.; De Martinis, E.C.P.; O’Brien, K.; Chen, Y.; Bradley, C.; et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome 2019, 7, 126. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Winglee, K.; Gharaibeh, R.Z.; Gauthier, J.; He, Z.; Tripathi, P.; Avram, D.; Bruner, S.; Fodor, A.; Jobin, C. Microbiota-Derived Metabolic Factors Reduce Campylobacteriosis in Mice. Gastroenterology 2018, 154, 1751–1763. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef]
- Sipe, L.M.; Chaib, M.; Pingili, A.K.; Pierre, J.F.; Makowski, L. Microbiome, bile acids, and obesity: How microbially modified metabolites shape anti-tumor immunity. Immunol. Rev. 2020, 295, 220–239. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Bjorck, I.; Backhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Villa, S.R.; Fuller, M.; Wicksteed, B.; Mackay, C.R.; Alquier, T.; Poitout, V.; Mancebo, H.; Mirmira, R.G.; Gilchrist, A.; et al. An Acetate-Specific GPCR, FFAR2, Regulates Insulin Secretion. Mol. Endocrinol. 2015, 29, 1055–1066. [Google Scholar] [CrossRef]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016, 24, 151–157. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.J.; Li, Y.R.; Cai, Z.M.; Li, S.H.; Zhu, J.F.; Zhang, F.; Liang, S.S.; Zhang, W.W.; Guan, Y.L.; Shen, D.Q.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Perez-Matute, P.; Marti, A.; Martinez, J.A.; Fernandez-Otero, M.P.; Stanhope, K.L.; Havel, P.J.; Moreno-Aliaga, M.J. Conjugated linoleic acid inhibits glucose metabolism, leptin and adiponectin secretion in primary cultured rat adipocytes. Mol. Cell Endocrinol. 2007, 268, 50–58. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.D. Banting lecture 2001: Dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes 2002, 51, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.H. Emerging Perspectives on Essential Amino Acid Metabolism in Obesity and the Insulin-Resistant State. Adv. Nutr. 2011, 2, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Frohnert, B.I.; Bernlohr, D.A. Protein Carbonylation, Mitochondrial Dysfunction, and Insulin Resistance. Adv. Nutr. 2013, 4, 157–163. [Google Scholar] [CrossRef]
- Li, X.; Xu, Z.L.; Lu, X.; Yang, X.H.; Yin, P.Y.; Kong, H.W.; Yu, Y.; Xu, G.W. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal. Chim. Acta 2009, 633, 257–262. [Google Scholar] [CrossRef]
- Menni, C.; Fauman, E.; Erte, I.; Perry, J.R.B.; Kastenmuller, G.; Shin, S.Y.; Petersen, A.K.; Hyde, C.; Psatha, M.; Ward, K.J.; et al. Biomarkers for Type 2 Diabetes and Impaired Fasting Glucose Using a Nontargeted Metabolomics Approach. Diabetes 2013, 62, 4270–4276. [Google Scholar] [CrossRef]
- Rhee, E.P.; Cheng, S.; Larson, M.G.; Walford, G.A.; Lewis, G.D.; McCabe, E.; Yang, E.; Farrell, L.; Fox, C.S.; O’Donnell, C.J.; et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Investig. 2011, 121, 1402–1411. [Google Scholar] [CrossRef]
- Walford, G.A.; Davis, J.; Warner, A.S.; Ackerman, R.J.; Billings, L.K.; Chamarthi, B.; Fanelli, R.R.; Hernandez, A.M.; Huang, C.; Khan, S.Q.; et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism 2013, 62, 1772–1778. [Google Scholar] [CrossRef]
- Wurtz, P.; Soininen, P.; Kangas, A.J.; Ronnemaa, T.; Lehtimaki, T.; Kahonen, M.; Viikari, J.S.; Raitakari, O.T.; Ala-Korpela, M. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 2013, 36, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [PubMed]

| Index | Control (n = 10) | T2DM (n = 7) | IGR (n = 5) |
|---|---|---|---|
| Age | 10.20 ± 4.26 | 12.58 ± 6.02 | 9.00 ± 2.19 |
| BMI | 15.05 ± 1.27 | 16.19 ± 2.00 | 14.2 ± 0.83 |
| FPG (mmol/L) | 4.10 ± 0.94 | 8.54 ± 1.12 ** | 6.50 ± 0.25 ## |
| HbA1c (%) | 3.32 ± 0.55 | 4.96 ± 2.50 * | 4.04 ± 0.45 # |
| FPI (μU/mL) | 5.97 ± 2.19 | 16.28 ± 8.89 ** | 11.87 ± 1.07 ## |
| IR | 1.11 ± 0.55 | 7.21 ± 3.29 **+ | 3.43 ± 0.32 ## |
| TG (mmol/L) | 0.37 ± 0.11 | 0.89 ± 0.76 | 0.71 ± 0.21 |
| TC (mmol/L) | 3.34 ± 0.80 | 3.46 ± 0.54 | 3.65 ± 0.99 |
| HDL (mmol/L) | 1.32 ± 0.35 | 1.36 ± 0.22 | 1.34 ± 0.46 |
| LDL (mmol/L) | 1.41 ± 0.51 | 1.35 ± 0.36 | 1.57 ± 0.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.; Pan, X.; Luo, J.; Liu, X.; Zhang, L.; Liu, Y.; Lei, G.; Hu, G.; Li, J. Alterations in Microbiota and Metabolites Related to Spontaneous Diabetes and Pre-Diabetes in Rhesus Macaques. Genes 2022, 13, 1513. https://doi.org/10.3390/genes13091513
Jiang C, Pan X, Luo J, Liu X, Zhang L, Liu Y, Lei G, Hu G, Li J. Alterations in Microbiota and Metabolites Related to Spontaneous Diabetes and Pre-Diabetes in Rhesus Macaques. Genes. 2022; 13(9):1513. https://doi.org/10.3390/genes13091513
Chicago/Turabian StyleJiang, Cong, Xuan Pan, Jinxia Luo, Xu Liu, Lin Zhang, Yun Liu, Guanglun Lei, Gang Hu, and Jing Li. 2022. "Alterations in Microbiota and Metabolites Related to Spontaneous Diabetes and Pre-Diabetes in Rhesus Macaques" Genes 13, no. 9: 1513. https://doi.org/10.3390/genes13091513
APA StyleJiang, C., Pan, X., Luo, J., Liu, X., Zhang, L., Liu, Y., Lei, G., Hu, G., & Li, J. (2022). Alterations in Microbiota and Metabolites Related to Spontaneous Diabetes and Pre-Diabetes in Rhesus Macaques. Genes, 13(9), 1513. https://doi.org/10.3390/genes13091513





