Genome-Wide Association Study of Airway Wall Thickening in a Korean Chronic Obstructive Pulmonary Disease Cohort
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Spirometry and Imaging Procedures
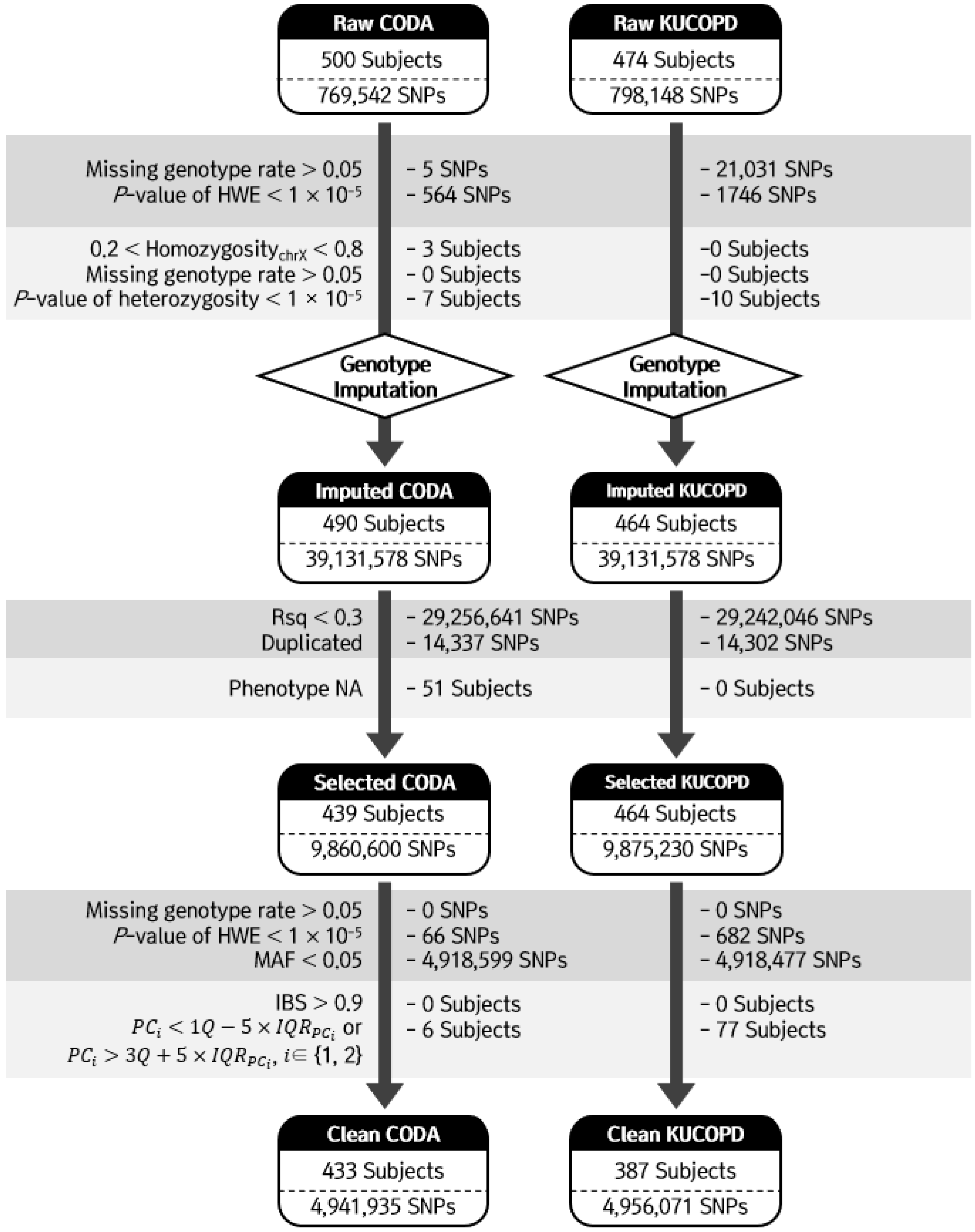
2.3. Genotyping and Quality Controls
2.4. Genotype Imputation of SNP Genotype Data
2.5. Meta-Analysis of GWAS
2.6. RNA Expression Association Test
3. Results
3.1. Subject Characteristics
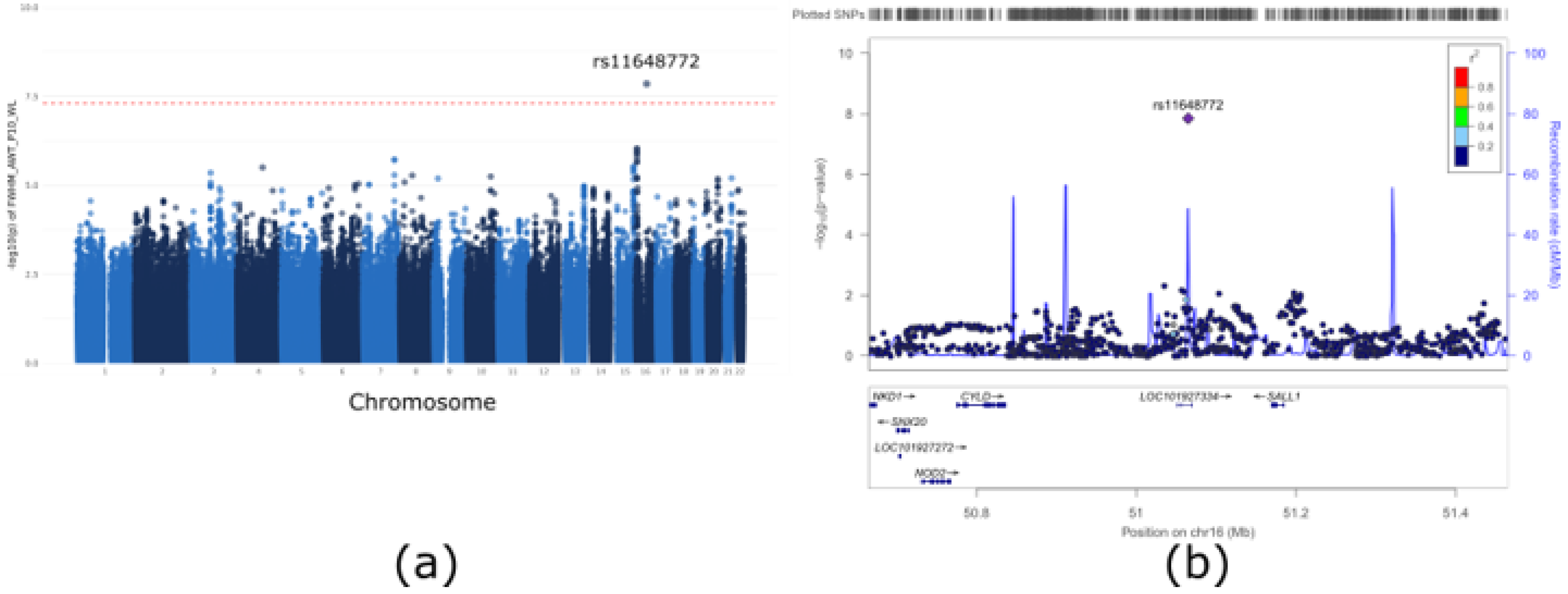
3.2. GWAS of Airway Wall Thickness
3.3. Analysis of Differentially Expressed Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bidan, C.M.; Veldsink, A.C.; Meurs, H.; Gosens, R. Airway and Extracellular Matrix Mechanics in COPD. Front. Physiol. 2015, 6, 346. [Google Scholar] [CrossRef] [PubMed]
- Şerifoğlu, İ.; Ulubay, G. The Methods Other than Spirometry in the Early Diagnosis of COPD. Tuberk. Toraks. 2019, 67, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.E.; Postma, D.S.; van Ginneken, B.; Wielputz, M.O.; Schmidt, M.; Becker, N.; Owsijewitsch, M.; Kauczor, H.U.; de Koning, H.J.; Lammers, J.W.; et al. Novel Genes for Airway Wall Thickness Identified with Combined Genome-Wide Association and Expression Analyses. Am. J. Respir. Crit. Care Med. 2014, 191, 547–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niimi, A.; Matsumoto, H.; Amitani, R.; Nakano, Y.; Mishima, M.; Minakuchi, M.; Nishimura, K.; Itoh, H.; Izumi, T. Airway Wall Thickness in Asthma Assessed by Computed Tomography. Relation to Clinical Indices. Am. J. Respir. Crit. Care Med. 2000, 162, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Mets, O.M.; de Jong, P.A.; van Ginneken, B.; Gietema, H.A.; Lammers, J.W.J. Quantitative Computed Tomography in COPD: Possibilities and Limitations. Lung 2012, 190, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, M.H.; Castaldi, P.J.; Hersh, C.P.; Hobbs, B.D.; Graham Barr, R.; Tal-Singer, R.; Bakke, P.; Gulsvik, A.; San José Estépar, R.; Van Beek, E.J.R. A Genome-Wide Association Study of Emphysema and Airway Quantitative Imaging Phenotypes. Am. J. Respir. Crit. Care Med. 2015, 192, 559–569. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Do, A.R.; Kang, H.Y.; Kim, W.J.; Lee, S.; Lee, J.H.; Song, W.J.; Kwon, H.S.; Cho, Y.S.; Moon, H.B.; et al. Genome-Wide Association Study of Korean Asthmatics: A Comparison With UK Asthmatics. Allergy Asthma Immunol. Res. 2021, 13, 609–622. [Google Scholar] [CrossRef]
- Han, Y.; Heo, Y.; Hong, Y.; Kwon, S.O.; Kim, W.J. Correlation between Physical Activity and Lung Function in Dusty Areas: Results from the Chronic Obstructive Pulmonary Disease in Dusty Areas (CODA) Cohort. Tuberc. Respir. Dis. 2019, 82, 311–318. [Google Scholar] [CrossRef]
- Shin, C.; Kim, J.; Kim, J.; Lee, S.; Shim, J.; In, K.; Kang, K.; Yoo, S.; Cho, N.; Kimm, K.; et al. Association of Habitual Snoring with Glucose and Insulin Metabolism in Nonobese Korean Adult Men. Am. J. Respir. Crit. Care Med. 2005, 171, 287–291. [Google Scholar] [CrossRef]
- Kim, S.; Lee, K.Y.; Kim, N.H.; Abbott, R.D.; Kim, C.; Lee, S.K.; Kim, S.H.; Shin, C. Relationship of Obstructive Sleep Apnoea Severity and Subclinical Systemic Atherosclerosis. Eur. Respir. J. 2020, 55, 1900959. [Google Scholar] [CrossRef]
- Miller, M.R.; Hankinson, J.; Brusasco, V.; Burgos, F.; Casaburi, R.; Coates, A.; Crapo, R.; Enright, P.; van der Grinten, C.P.M.; Gustafsson, P.; et al. Standardisation of Spirometry. Eur. Respir. J. 2005, 26, 319–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakkar, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.E.; Lee, S.; Park, K.; Elston, R.C.; Yang, H.J.; Won, S. ONETOOL for the Analysis of Family-Based Big Data. Bioinformatics 2018, 34, 2851–2853. [Google Scholar] [CrossRef] [PubMed]
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A Note on Exact Tests of Hardy-Weinberg Equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Haplotype Reference Consortium. A Reference Panel of 64,976 Haplotypes for Genotype Imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-Based Phasing Using the Haplotype Reference Consortium Panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-Generation Genotype Imputation Service and Methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Kicic, A.; Hallstrand, T.S.; Sutanto, E.N.; Stevens, P.T.; Kobor, M.S.; Taplin, C.; Paré, P.D.; Beyer, R.P.; Stick, S.M.; Knight, D.A. Decreased Fibronectin Production Significantly Contributes to Dysregulated Repair of Asthmatic Epithelium. Am. J. Respir. Crit. Care Med. 2010, 181, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Park, K.; An, J.; Gim, J.; Seo, M.; Lee, W.; Park, T.; Won, S. BALLI: Bartlett-Adjusted Likelihood-Based Linear Model Approach for Identifying Differentially Expressed Genes with RNA-Seq Data. BMC Genom. 2019, 20, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, S.M.; Cho, M.H.; Young, K.; Hersh, C.P.; Castaldi, P.J.; McDonald, M.; Regan, E.; Mattheisen, M.; DeMeo, D.L.; Parker, M.; et al. A Genome-Wide Association Study Identifies Risk Loci for Spirometric Measures Among Smokers of European and African Ancestry. BMC Genet. 2015, 16, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benlala, I.; Laurent, F.; Dournes, G. Structural and Functional Changes in COPD: What We Have Learned from Imaging. Respirology 2021, 26, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Cho, M.H.; Castaldi, P.J.; Hersh, C.P.; Silverman, E.K.; Laird, N.M. Heritability of Chronic Obstructive Pulmonary Disease and Related Phenotypes in Smokers. Am. J. Respir. Crit. Care Med. 2013, 188, 941–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, D.; Zhang, W.; Jia, Y.; Cong, D.; Hu, S. Spalt-Like Transcription Factor 1 (SALL1) Gene Expression Inhibits Cell Proliferation and Cell Migration of Human Glioma Cells Through the Wnt/β-Catenin Signaling Pathway. Med. Sci. Monit. Basic Res. 2019, 25, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Zou, Y.; Zhao, Z.; Li, B.; Ran, P. Nicotine-Induced Epithelial-Mesenchymal Transition via Wnt/ β-Catenin Signaling in Human Airway Epithelial Cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L199–L209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohal, S.S.; Reid, D.; Soltani, A.; Ward, C.; Weston, S.; Muller, H.K.; Wood-Baker, R.; Walters, E.H. Reticular Basement Membrane Fragmentation and Potential Epithelial Mesenchymal Transition is Exaggerated in the Airways of Smokers with Chronic Obstructive Pulmonary Disease. Respirology 2010, 15, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Kanellopoulos, J.M.; Ojcius, D.M. Neither B Cell Nor T Cell—The Unique Group of Innate Lymphoid Cells. Biomed. J. 2021, 44, 112–114. [Google Scholar] [CrossRef]
- Fonseca, W.; Lukacs, N.W.; Elesela, S.; Malinczak, C.A. Role of ILC2 in Viral-Induced Lung Pathogenesis. Front. Immunol. 2021, 12, 675169. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Luo, D.; Wang, X.; Zhang, Y.; Liu, Z.; Zhong, N.; Wu, M.; Li, G. Lyn Regulates Mucus Secretion and MUC5AC via the STAT6 Signaling Pathway During Allergic Airway Inflammation. Sci. Rep. 2017, 7, 42675. [Google Scholar] [CrossRef] [Green Version]

| Variable | CODA Cohort (n = 433) | KUCOPD Cohort (n = 387) | Total (n = 820) | p Value |
|---|---|---|---|---|
| Female sex, n (%) | 118 (27.3%) | 96 (24.8%) | 214 (26.1%) | 0.4737 |
| Age | 72.0 ± 7.1 | 63.0 ± 7.6 | 67.7 ± 8.6 | <0.001 |
| Height | 159.3 ± 9.3 | 164.1 ± 7.7 | 161.5 ± 8.9 | <0.001 |
| Weight | 59.9 ± 10.2 | 65.7 ± 9.5 | 62.7 ± 10.3 | <0.001 |
| BMI | 23.6 ± 3.2 | 24.4 ± 2.9 | 24.0 ± 3.1 | <0.001 |
| Pack-Year | 17.3 ± 22.9 | 16.6 ± 20.3 | 17.0 ± 21.7 | 0.8058 |
| Smoking, n (%) | 0.4748 | |||
| Never | 156 (36.2%) | 152 (39.4%) | 308 (37.7%) | |
| Former | 179 (41.5%) | 160 (41.5%) | 339 (41.5%) | |
| Current | 96 (22.3%) | 74 (19.2%) | 170 (20.8%) | |
| FEV1. Pred. pre-BD (%) | 83.9 ± 22.7 | 101.7 ± 17.0 | 92.0 ± 22.1 | <0.001 |
| FEV/FVC (%) | 65.1 ± 11.5 | 71.2 ± 8.7 | 67.9 ± 10.7 | <0.001 |
| COPD, n (%) | 278 (64.2%) | 194 (50.1%) | 472 (57.6%) | <0.001 |
| FWHM_AWT_P10_WL | 4.7 ± 0.4 | 4.4 ± 0.2 | 4.5 ± 0.3 | <0.001 |
| Chr | BP | SNP | MAFKRef | Effect | SE | p Value | Alt /Ref | CODA Cohort | KUCOPD Cohort | Gene | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p Value | MAF | HWE | p Value | MAF | HWE | |||||||||
| 16 | 51065076 | rs11648772 | 0.19 | −0.08 | 0.01 | 1.41 × 10−8 | G/T | 0.0240 | 0.24 | 0.6950 | 2.87 × 10−7 | 0.25 | 0.4168 | LINC02127 |
| 16 | 9109561 | rs12931044 | 0.13 | 0.09 | 0.02 | 9.22 × 10−7 | A/G | 0.2466 | 0.13 | 1.0000 | 1.05 × 10−6 | 0.13 | 0.5096 | USP7 (dist = 52,220), C16orf72 (dist = 75,976) |
| 7 | 139176178 | rs11970854 | 0.45 | 0.06 | 0.01 | 1.92 × 10−6 | T/C | 0.0012 | 0.47 | 0.9235 | 3.35 × 10−4 | 0.49 | 0.9190 | KLRG2 (dist = 7721), CLEC2L (dist = 32,496) |
| 15 | 97215196 | rs4383104 | 0.10 | 0.09 | 0.02 | 3.02 × 10−6 | G/A | 0.0006 | 0.12 | 0.2442 | 8.28 × 10−4 | 0.09 | 0.3565 | NR2F2 (dist = 331,704), SPATA8-AS1 (dist = 100,039) |
| 4 | 113212158 | rs150827063 | 0.10 | 0.11 | 0.02 | 3.15 × 10−6 | A/G | 0.7282 | 0.09 | 0.7667 | 2.07 × 10−7 | 0.07 | 0.7092 | TIFA (dist = 5099), ALPK1 (dist = 6341) |
| 3 | 87115233 | rs56694856 | 0.14 | 0.08 | 0.02 | 4.45 × 10−6 | T/C | 0.0015 | 0.15 | 0.5605 | 5.12 × 10−4 | 0.17 | 0.5816 | VGLL3 (dist = 74,960), LINC00506 (dist = 23,197) |
| 8 | 56895276 | rs16920168 | 0.10 | 0.09 | 0.02 | 5.29 × 10−6 | T/C | 0.0127 | 0.10 | 0.2902 | 1.58 × 10−4 | 0.09 | 0.3644 | LYN |
| 10 | 107851568 | rs673353 | 0.33 | −0.06 | 0.01 | 5.68 × 10−6 | G/T | 0.8980 | 0.33 | 0.8277 | 6.05 × 10−7 | 0.33 | 0.2038 | LOC101927549 (dist = 271,477), LOC105378470 (dist = 48,355) |
| 21 | 45757631 | rs56242109 | 0.13 | −0.09 | 0.02 | 6.27 × 10−6 | A/G | 0.0687 | 0.12 | 0.5001 | 3.77 × 10−5 | 0.12 | 0.0962 | C21orf2 |
| 9 | 21359818 | rs140373339 | 0.03 | 0.11 | 0.03 | 6.41 × 10−6 | A/C | 0.0220 | 0.06 | 0.6320 | 1.18 × 10−4 | 0.06 | 0.1563 | IFNA6 (dist = 8932), IFNA13 (dist = 7553) |
| Gene | Chr | BP | Coefficient | p Value | ||
|---|---|---|---|---|---|---|
| limma | balli | limma | balli | |||
| SALL1 | 16 | 51,169,886–51,185,278 | 0.12 | 0.12 | 0.0173 | 0.0092 |
| CYLD | 16 | 50,775,961–50,835,846 | 0.02 | 0.84 | 0.8502 | 0.9079 |
| NOD2 | 16 | 50,727,514–50,766,988 | 0.03 | 0.64 | 0.6576 | 0.7697 |
| BRD7 | 16 | 50,347,398–50,402,845 | 0.01 | 0.93 | 0.9312 | 0.9609 |
| ADCY7 | 16 | 50,280,048–50,352,046 | −0.01 | 0.93 | 0.9764 | 0.9864 |
| HEATR3 | 16 | 50,099,852–50,140,298 | 0.11 | 0.33 | 0.3587 | 0.5156 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, A.R.; Ko, D.Y.; Kim, J.; Bak, S.H.; Lee, K.Y.; Yoon, D.; Shin, C.; Kim, S.; Kim, W.J.; Won, S. Genome-Wide Association Study of Airway Wall Thickening in a Korean Chronic Obstructive Pulmonary Disease Cohort. Genes 2022, 13, 1258. https://doi.org/10.3390/genes13071258
Do AR, Ko DY, Kim J, Bak SH, Lee KY, Yoon D, Shin C, Kim S, Kim WJ, Won S. Genome-Wide Association Study of Airway Wall Thickening in a Korean Chronic Obstructive Pulmonary Disease Cohort. Genes. 2022; 13(7):1258. https://doi.org/10.3390/genes13071258
Chicago/Turabian StyleDo, Ah Ra, Do Yeon Ko, Jeeyoung Kim, So Hyeon Bak, Ki Yeol Lee, Dankyu Yoon, Chol Shin, Soriul Kim, Woo Jin Kim, and Sungho Won. 2022. "Genome-Wide Association Study of Airway Wall Thickening in a Korean Chronic Obstructive Pulmonary Disease Cohort" Genes 13, no. 7: 1258. https://doi.org/10.3390/genes13071258
APA StyleDo, A. R., Ko, D. Y., Kim, J., Bak, S. H., Lee, K. Y., Yoon, D., Shin, C., Kim, S., Kim, W. J., & Won, S. (2022). Genome-Wide Association Study of Airway Wall Thickening in a Korean Chronic Obstructive Pulmonary Disease Cohort. Genes, 13(7), 1258. https://doi.org/10.3390/genes13071258







