Drug-Target Network Study Reveals the Core Target-Protein Interactions of Various COVID-19 Treatments
Abstract
:1. Introduction
2. Materials and Methods
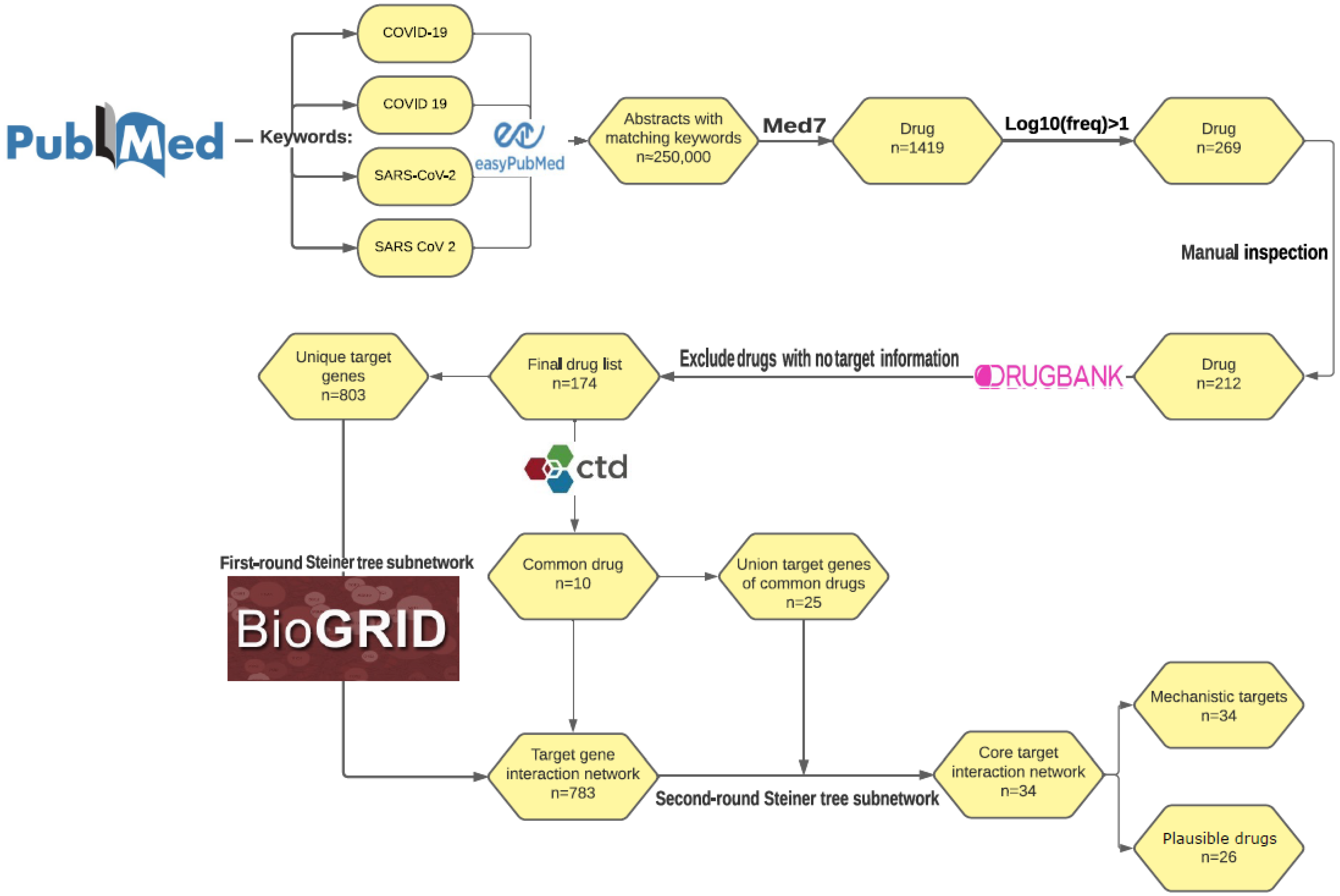
2.1. Text Mining of COVID-19 Drugs and Drug Target Curation
2.2. Steiner Tree Analysis
2.3. Functional Enrichment Analysis
2.4. GWAS Summary Statistics Process and z-Score Permutation
2.5. Differentially Expressed Gene Analysis for COVID-19 Single-Cell RNA-Seq Data
3. Results
3.1. Identifying COVID-19 Drugs and Corresponding Targets via Literature Mining and Curation
3.2. COVID-19 Drug Target-Protein Network
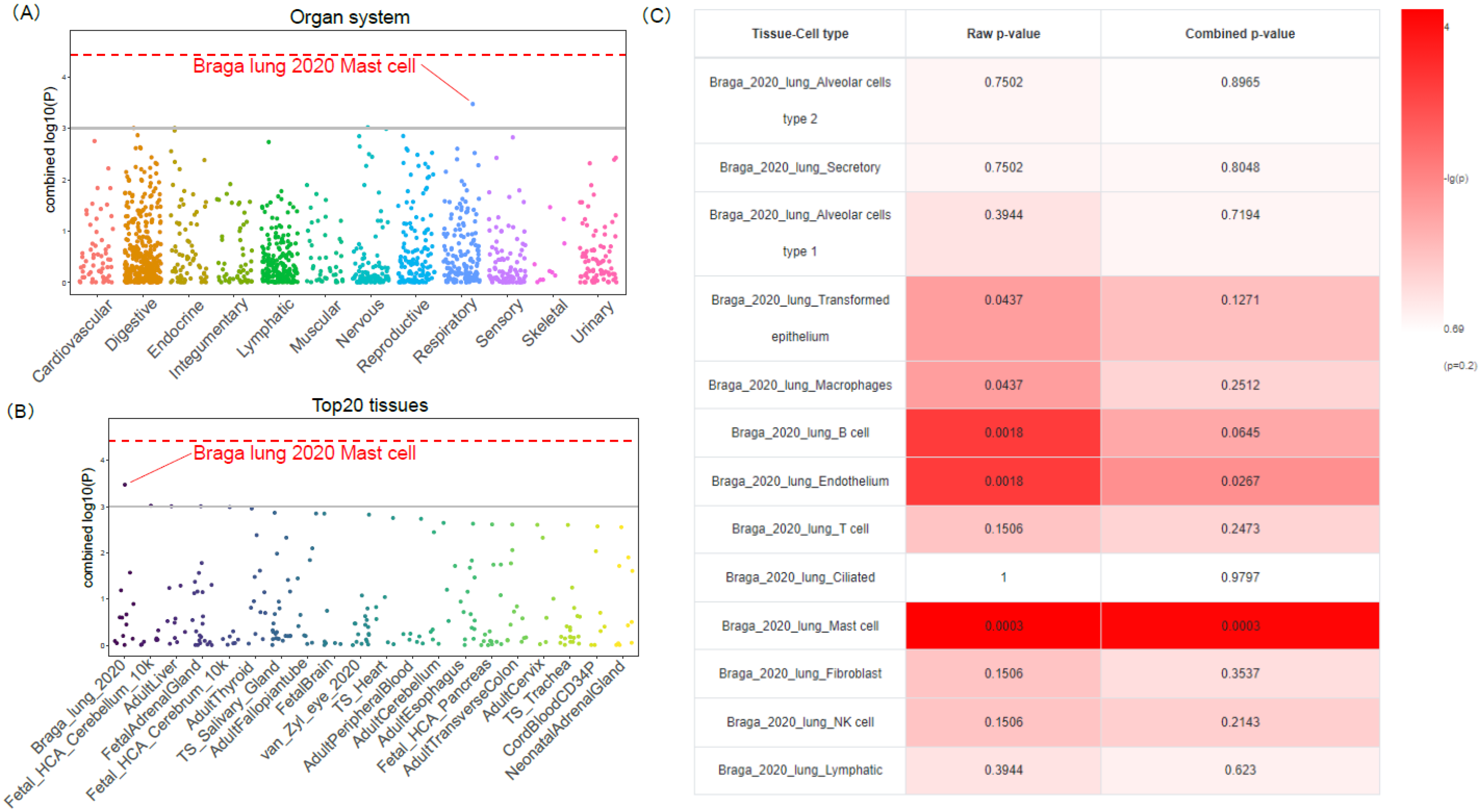
3.3. Functional Enrichment and Tissue-Cell-Type Specificity of COVID19-DrugNET Genes
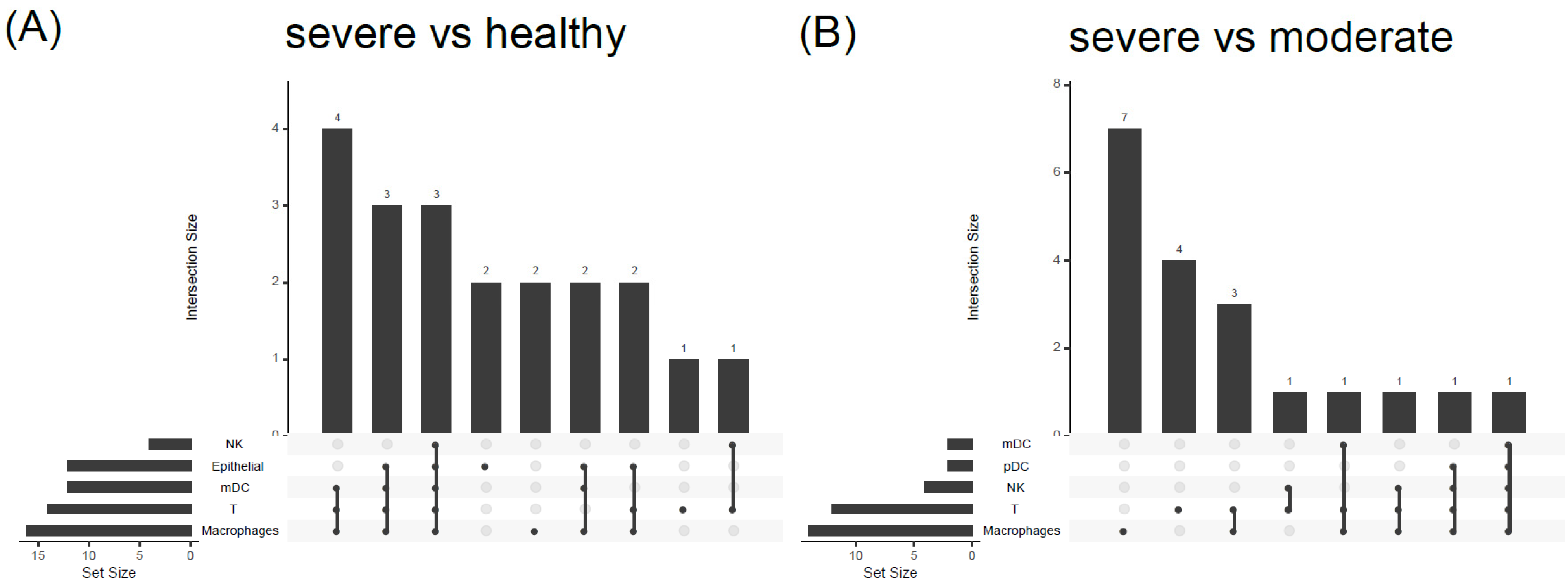
3.4. COVID19-DrugNET Is Highly Related to Risk Genes Underlying Severe COVID-19 Symptoms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. COVID-19 Weekly Epidemiological Update, Edition 80, 22 February 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Ye, Z.W.; Yuan, S.; Yuen, K.S.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Remdesivir: First approval. Drugs 2020, 80, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. COVID-19: Pfizer’s Paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. Molnupiravir in COVID-19: A systematic review of literature. Diabetes Metab. Syndr. 2021, 15, 102329. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.C.; Lu, R.M.; Su, S.C.; Chiang, P.Y.; Ko, S.H.; Ke, F.Y.; Liang, K.H.; Hsieh, T.Y.; Wu, H.C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed Sci. 2022, 29, 1. [Google Scholar] [CrossRef]
- Vicenti, I.; Zazzi, M.; Saladini, F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021, 31, 325–337. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, J.Y.; Yang, J.W.; Lee, K.H.; Effenberger, M.; Szpirt, W.; Kronbichler, A.; Shin, J.I. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics 2021, 11, 316–329. [Google Scholar] [CrossRef]
- Israel, A.; Schaffer, A.A.; Cicurel, A.; Cheng, K.; Sinha, S.; Schiff, E.; Feldhamer, I.; Tal, A.; Lavie, G.; Ruppin, E. Identification of drugs associated with reduced severity of COVID-19—A case-control study in a large population. eLife 2021, 10, e68165. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, C.E.; Pinsky, B.A.; Brogdon, J.; Chen, C.; Ruder, K.; Zhong, L.; Nyatigo, F.; Cook, C.A.; Hinterwirth, A.; Lebas, E.; et al. Effect of oral azithromycin vs. placebo on COVID-19 symptoms in outpatients with SARS-CoV-2 infection: A randomized clinical trial. JAMA 2021, 326, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Wang, W.; Hayek, S.S.; Chan, L.; Mathews, K.S.; Melamed, M.L.; Brenner, S.K.; Leonberg-Yoo, A.; Schenck, E.J.; Radbel, J.; et al. Association between early treatment with tocilizumab and mortality among critically ill patients with COVID-19. JAMA Intern. Med. 2021, 181, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Offit, P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA 2021, 325, 821–822. [Google Scholar] [CrossRef]
- Excler, J.L.; Saville, M.; Berkley, S.; Kim, J.H. Vaccine development for emerging infectious diseases. Nat. Med. 2021, 27, 591–600. [Google Scholar] [CrossRef]
- Schenone, M.; Dancik, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, C.; Zitnik, M.; Leskovec, J. Identification of disease treatment mechanisms through the multiscale interactome. Nat. Commun. 2021, 12, 1796. [Google Scholar] [CrossRef]
- Yildirim, M.A.; Goh, K.I.; Cusick, M.E.; Barabasi, A.L.; Vidal, M. Drug-target network. Nat. Biotechnol. 2007, 25, 1119–1126. [Google Scholar] [CrossRef]
- Casas, A.I.; Hassan, A.A.; Larsen, S.J.; Gomez-Rangel, V.; Elbatreek, M.; Kleikers, P.W.M.; Guney, E.; Egea, J.; Lopez, M.G.; Baumbach, J.; et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl. Acad. Sci. USA 2019, 116, 7129–7136. [Google Scholar] [CrossRef] [Green Version]
- Sakle, N.S.; More, S.A.; Mokale, S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Sci. Rep. 2020, 10, 17217. [Google Scholar] [CrossRef]
- Wu, Z.; Li, W.; Liu, G.; Tang, Y. Network-based methods for prediction of drug-target interactions. Front. Pharmacol. 2018, 9, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recanatini, M.; Cabrelle, C. Drug research meets network science: Where are we? J. Med. Chem. 2020, 63, 8653–8666. [Google Scholar] [CrossRef] [PubMed]
- Manuel, A.M.; Dai, Y.; Freeman, L.A.; Jia, P.; Zhao, Z. An integrative study of genetic variants with brain tissue expression identifies viral etiology and potential drug targets of multiple sclerosis. Mol. Cell Neurosci. 2021, 115, 103656. [Google Scholar] [CrossRef]
- Bagga, S.; Bouchard, M.J. Cell cycle regulation during viral infection. Methods Mol. Biol. 2014, 1170, 165–227. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Seth, S.; Batra, J.; Srinivasan, S. COVID-19: Targeting Proteases in Viral Invasion and Host Immune Response. Front. Mol. Biosci. 2020, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Fajgenbaum, D.C. Is severe COVID-19 a cytokine storm syndrome: A hyperinflammatory debate. Curr. Opin. Rheumatol. 2021, 33, 419–430. [Google Scholar] [CrossRef]
- Zambrana, C.; Xenos, A.; Bottcher, R.; Malod-Dognin, N.; Przulj, N. Network neighbors of viral targets and differentially expressed genes in COVID-19 are drug target candidates. Sci. Rep. 2021, 11, 18985. [Google Scholar] [CrossRef]
- Hsieh, K.L.; Wang, Y.; Chen, L.; Zhao, Z.; Savitz, S.; Jiang, X.; Tang, J.; Kim, Y. Drug repurposing for COVID-19 using graph neural network and harmonizing multiple evidence. Sci. Rep. 2021, 11, 23179. [Google Scholar] [CrossRef]
- Kormilitzin, A.; Vaci, N.; Liu, Q.; Nevado-Holgado, A. Med7: A transferable clinical natural language processing model for electronic health records. Artif. Intell. Med. 2021, 118, 102086. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.; Ravi, R. A Nearly Best-Possible Approximation Algorithm for Node-Weighted Steiner Trees. J. Algorithms 1995, 19, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.S.; Perkins, T.; Bunnell, S.; Pepin, F.; Thomas, D.Y.; Hallett, M. Identifying regulatory subnetworks for a set of genes. Mol. Cell Proteom. 2005, 4, 683–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxon, J.A.; Yu, H.; Polosukhin, V.V.; Stathopoulos, G.T.; Gleaves, L.A.; McLoed, A.G.; Massion, P.P.; Yull, F.E.; Zhao, Z.; Blackwell, T.S. p52 expression enhances lung cancer progression. Sci. Rep. 2018, 8, 6078. [Google Scholar] [CrossRef] [Green Version]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pei, G.; Yan, F.; Simon, L.M.; Dai, Y.; Jia, P.; Zhao, Z. deCS: A tool for systematic cell type annotations of single-cell RNA sequencing data among human tissues. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, R.; Liu, A.; Cho, K.S.; Manuel, A.M.; Li, X.; Dong, X.; Jia, P.; Zhao, Z. WebCSEA: Web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 2022, 50, W782–W790. [Google Scholar] [CrossRef]
- Pei, G.; Dai, Y.; Zhao, Z.; Jia, P. deTS: Tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics 2019, 35, 3842–3845. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, R.; Manuel, A.M.; Liu, A.; Jia, P.; Zhao, Z. CSEA-DB: An omnibus for human complex trait and cell type associations. Nucleic Acids Res. 2021, 49, D862–D870. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Dai, Y.; Hu, R.; Pei, G.; Manuel, A.M.; Zhao, Z. TSEA-DB: A trait-tissue association map for human complex traits and diseases. Nucleic Acids Res. 2020, 48, D1022–D1030. [Google Scholar] [CrossRef] [PubMed]
- Severe Covid-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; et al. Genomewide association study of severe COVID-19 with respiratory failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. Available online: https://www.covid19hg.org/ (accessed on 23 August 2020).
- National Institute of Health. COVID-19 GWAS Results—for Public Download/Use. Available online: https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx (accessed on 1 March 2022).
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive integration of single-cell data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef]
- Southan, C.; Sitzmann, M.; Muresan, S. Comparing the chemical structure and protein content of ChEMBL, DrugBank, Human Metabolome Database and the Therapeutic Target Database. Mol. Inform. 2013, 32, 881–897. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, R.; Zhao, Z. WebCSEA: Web-Based Cell-type Specific Enrichment Analysis of Genes. Available online: https://bioinfo.uth.edu/webcsea/ (accessed on 5 February 2022).
- Motta Junior, J.D.S.; Miggiolaro, A.; Nagashima, S.; de Paula, C.B.V.; Baena, C.P.; Scharfstein, J.; de Noronha, L. Mast cells in alveolar septa of COVID-19 patients: A pathogenic pathway that may link interstitial edema to immunothrombosis. Front. Immunol. 2020, 11, 574862. [Google Scholar] [CrossRef] [PubMed]
- Gebremeskel, S.; Schanin, J.; Coyle, K.M.; Butuci, M.; Luu, T.; Brock, E.C.; Xu, A.; Wong, A.; Leung, J.; Korver, W.; et al. Mast cell and eosinophil activation are associated with COVID-19 and TLR-mediated viral inflammation: Implications for an anti-siglec-8 antibody. Front. Immunol. 2021, 12, 650331. [Google Scholar] [CrossRef] [PubMed]
- Initiative, C.-H.G. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, J.; Jeong, H.H.; Chen, W.; Jia, P.; Zhao, Z. Association of CXCR6 with COVID-19 severity: Delineating the host genetic factors in transcriptomic regulation. Hum. Genet. 2021, 140, 1313–1328. [Google Scholar] [CrossRef]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 12, 720109. [Google Scholar] [CrossRef]
- Angelini, J.; Talotta, R.; Roncato, R.; Fornasier, G.; Barbiero, G.; Dal Cin, L.; Brancati, S.; Scaglione, F. JAK-inhibitors for the treatment of rheumatoid arthritis: A focus on the present and an outlook on the future. Biomolecules 2020, 10, 1002. [Google Scholar] [CrossRef]
- Wang, X.; Wen, Y.; Xie, X.; Liu, Y.; Tan, X.; Cai, Q.; Zhang, Y.; Cheng, L.; Xu, G.; Zhang, S.; et al. Dysregulated hematopoiesis in bone marrow marks severe COVID-19. Cell Discov. 2021, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Bruton tyrosine kinase inhibitors: Present and future. Cancer J. 2019, 25, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Kosiborod, M.N.; Esterline, R.; Furtado, R.H.M.; Oscarsson, J.; Gasparyan, S.B.; Koch, G.G.; Martinez, F.; Mukhtar, O.; Verma, S.; Chopra, V.; et al. Dapagliflozin in patients with cardiometabolic risk factors hospitalised with COVID-19 (DARE-19): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Diabetes Endocrinol. 2021, 9, 586–594. [Google Scholar] [CrossRef]
- Iovino, L.; Thur, L.A.; Gnjatic, S.; Chapuis, A.; Milano, F.; Hill, J.A. Shared inflammatory pathways and therapeutic strategies in COVID-19 and cancer immunotherapy. J. Immunother. Cancer 2021, 9, e002392. [Google Scholar] [CrossRef]
- Vergoten, G.; Bailly, C. Interaction of the renin inhibitor aliskiren with the SARS-CoV-2 main protease: A molecular docking study. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aliter, K.F.; Al-Horani, R.A. Thrombin inhibition by argatroban: Potential therapeutic benefits in COVID-19. Cardiovasc. Drugs Ther. 2021, 35, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Welen, K.; Rosendal, E.; Gisslen, M.; Lenman, A.; Freyhult, E.; Fonseca-Rodriguez, O.; Bremell, D.; Stranne, J.; Balkhed, A.O.; Niward, K.; et al. A phase 2 trial of the effect of antiandrogen therapy on COVID-19 outcome: No evidence of benefit, supported by epidemiology and in vitro data. Eur. Urol. 2021, 81, 285–293. [Google Scholar] [CrossRef]
- Maynard, S.; Ros-Soto, J.; Chaidos, A.; Innes, A.; Paleja, K.; Mirvis, E.; Buti, N.; Sharp, H.; Palanicawandar, R.; Milojkovic, D. The role of ibrutinib in COVID-19 hyperinflammation: A case report. Int. J. Infect. Dis. 2021, 105, 274–276. [Google Scholar] [CrossRef]
- Fidan, C.; Aydogdu, A. As a potential treatment of COVID-19: Montelukast. Med. Hypotheses 2020, 142, 109828. [Google Scholar] [CrossRef] [PubMed]
- Akinbolade, S.; Coughlan, D.; Fairbairn, R.; McConkey, G.; Powell, H.; Ogunbayo, D.; Craig, D. Combination therapies for COVID-19: An overview of the clinical trials landscape. Br. J. Clin. Pharmacol. 2022, 88, 1590–1597. [Google Scholar] [CrossRef] [PubMed]
- Gavriatopoulou, M.; Ntanasis-Stathopoulos, I.; Korompoki, E.; Fotiou, D.; Migkou, M.; Tzanninis, I.G.; Psaltopoulou, T.; Kastritis, E.; Terpos, E.; Dimopoulos, M.A. Emerging treatment strategies for COVID-19 infection. Clin. Exp. Med. 2021, 21, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus remdesivir for hospitalized adults with COVID-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef]

| Gene Symbol | Degree | Betweenness |
|---|---|---|
| HSP90AA1 | 196 | 37,757.90 |
| TP53 | 148 | 17,451.41 |
| APP | 145 | 32,447.83 |
| NTRK1 | 141 | 15,363.83 |
| MYC | 134 | 12,058.01 |
| EGFR | 125 | 19,208.06 |
| ESR1 | 114 | 7602.044 |
| ESR2 | 89 | 8696.631 |
| EGLN3 | 83 | 5614.034 |
| XPO1 | 82 | 4984.01 |
| Drug | Compilation | CTDbase | Overlapping | Union | PharmGKB Annotation |
|---|---|---|---|---|---|
| acalabrutinib † | 1 | 3 | 0 | 4 | other |
| aliskiren | 1 | 3 | 0 | 4 | NA |
| Argatroban ‡ | 1 | 1 | 0 | 2 | NA |
| baricitinib †‡ | 4 | 11 | 3 | 12 | anti-cytokine/anti-inflammatory |
| bicalutamide | 1 | 4 | 0 | 5 | NA |
| dapagliflozin †‡ | 1 | 3 | 0 | 4 | other |
| Ibrutinib † | 1 | 4 | 1 | 4 | NA |
| montelukast †‡ | 2 | 6 | 0 | 8 | NA |
| ruxolitinib † | 4 | 4 | 0 | 8 | other |
| tofacitinib †‡ | 4 | 8 | 0 | 12 | anti-cytokine/anti-inflammatory |
| Drug | p.hyper | Target Reservation Rate | Reserved Target Genes |
|---|---|---|---|
| adalimumab | 0 | 1/1 | TNF |
| bromhexine | 0.0018 | 1/2 | TMPRSS2 |
| canakinumab | 0 | 1/1 | IL1B |
| deferoxamine | 0 | 1/1 | APP |
| epinephrine | 0.0036 | 2/8 | ADRB2, TNF |
| formoterol | 0.0053 | 1/3 | ADRB2 |
| infliximab | 0 | 1/1 | TNF |
| leflunomide | 0.0053 | 1/3 | PTK2B |
| progesterone | 0.0073 | 2/10 | ESR1, AR |
| tocilizumab | 0 | 1/1 | IL6R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Yu, H.; Yan, Q.; Li, B.; Liu, A.; Liu, W.; Jiang, X.; Kim, Y.; Guo, Y.; Zhao, Z. Drug-Target Network Study Reveals the Core Target-Protein Interactions of Various COVID-19 Treatments. Genes 2022, 13, 1210. https://doi.org/10.3390/genes13071210
Dai Y, Yu H, Yan Q, Li B, Liu A, Liu W, Jiang X, Kim Y, Guo Y, Zhao Z. Drug-Target Network Study Reveals the Core Target-Protein Interactions of Various COVID-19 Treatments. Genes. 2022; 13(7):1210. https://doi.org/10.3390/genes13071210
Chicago/Turabian StyleDai, Yulin, Hui Yu, Qiheng Yan, Bingrui Li, Andi Liu, Wendao Liu, Xiaoqian Jiang, Yejin Kim, Yan Guo, and Zhongming Zhao. 2022. "Drug-Target Network Study Reveals the Core Target-Protein Interactions of Various COVID-19 Treatments" Genes 13, no. 7: 1210. https://doi.org/10.3390/genes13071210
APA StyleDai, Y., Yu, H., Yan, Q., Li, B., Liu, A., Liu, W., Jiang, X., Kim, Y., Guo, Y., & Zhao, Z. (2022). Drug-Target Network Study Reveals the Core Target-Protein Interactions of Various COVID-19 Treatments. Genes, 13(7), 1210. https://doi.org/10.3390/genes13071210







