Molecular Basis of Pathogenic Variants in the Fibrillar Collagens
Abstract
1. Introduction
2. Biosynthesis of the Fibrillar Collagens
3. Pathogenic Changes
3.1. Glycine Substitutions
3.2. X and Y Position Substitutions
3.3. Null Alleles
3.4. Abnormal Splicing
3.5. In-Frame Insertions and Deletions
3.6. C-Propeptide Changes
3.7. N-Propeptide and Signal Peptide Sequence Changes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Prockop, D.J.; Constantinou, C.D.; Dombrowski, K.E.; Hojima, Y.; Kadler, K.E.; Kuivaniemi, H.; Tromp, G.; Vogel, B.E. Type I procollagen: The gene-protein system that harbors most of the mutations causing osteogenesis imperfecta and probably more common heritable disorders of connective tissue. Am. J. Med. Genet. 1989, 34, 60–67. [Google Scholar] [CrossRef]
- Myllyharju, J.; Kivirikko, K.I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004, 20, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Bateman, J.F.; Boot-Handford, R.P.; Lamandé, S.R. Genetic diseases of connective tissues: Cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009, 10, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Exposito, J.Y.; Valcourt, U.; Cluzel, C.; Lethias, C. The fibrillar collagen family. Int. J. Mol. Sci. 2010, 11, 407–426. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Birk, D.E. Focus on molecules: Collagens V and XI. Exp. Eye Res. 2012, 98, 105–106. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.F.; Tasab, M.; Bulleid, N.J. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 1997, 16, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Bächinger, H.P. A molecular ensemble in the rER for procollagen maturation. Biochim. Biophys. Acta 2013, 1833, 2479–2491. [Google Scholar] [CrossRef]
- Moradi-Améli, M.; Rousseau, J.C.; Kleman, J.P.; Champliaud, M.F.; Boutillon, M.M.; Bernillon, J.; Wallach, J.; Van der Rest, M. Diversity in the processing events at the N-terminus of type-V collagen. Eur. J. Biochem. 1994, 221, 987–995. [Google Scholar] [CrossRef]
- Rousseau, J.C.; Farjanel, J.; Boutillon, M.M.; Hartmann, D.J.; van der Rest, M.; Moradi-Améli, M. Processing of type XI collagen. Determination of the matrix forms of the alpha1(XI) chain. J. Biol. Chem. 1996, 271, 23743–23748. [Google Scholar] [CrossRef]
- Burgeson, R.E.; Hollister, D.W. Collagen heterogeneity in human cartilage: Identification of several new collagen chains. Biochem. Biophys. Res. Commun. 1979, 87, 1124–1131. [Google Scholar] [CrossRef]
- Reese, C.A.; Mayne, R. Minor collagens of chicken hyaline cartilage. Biochemistry 1981, 20, 5443–5448. [Google Scholar] [CrossRef] [PubMed]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.R.; Paz, M.A.; Gallop, P.M. Cross-linking in collagen and elastin. Annu. Rev. Biochem. 1984, 53, 717–748. [Google Scholar] [CrossRef] [PubMed]
- Kadler, K.E.; Hill, A.; Canty-Laird, E.G. Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 2008, 20, 495–501. [Google Scholar] [CrossRef]
- Revell, C.K.; Jensen, O.E.; Shearer, T.; Lu, Y.; Holmes, D.F.; Kadler, K.E. Collagen fibril assembly: New approaches to unanswered questions. Matrix Biol. Plus 2021, 12, 100079. [Google Scholar] [CrossRef]
- Musiime, M.; Chang, J.; Hansen, U.; Kadler, K.E.; Zeltz, C.; Gullberg, D. Collagen Assembly at the Cell Surface: Dogmas Revisited. Cells 2021, 10, 662. [Google Scholar] [CrossRef]
- Pope, F.M.; Martin, G.R.; Lichtenstein, J.R.; Penttinen, R.; Gerson, B.; Rowe, D.W.; McKusick, V.A. Patients with Ehlers-Danlos syndrome type IV lack type III collagen. Proc. Natl. Acad. Sci. USA 1975, 72, 1314–1316. [Google Scholar] [CrossRef]
- Bateman, J.F.; Mascara, T.; Chan, D.; Cole, W.G. Abnormal type I collagen metabolism by cultured fibroblasts in lethal perinatal osteogenesis imperfecta. Biochem. J. 1984, 217, 103–115. [Google Scholar] [CrossRef]
- Steinmann, B.; Rao, V.H.; Vogel, A.; Bruckner, P.; Gitzelmann, R.; Byers, P.H. Cysteine in the triple-helical domain of one allelic product of the alpha 1(I) gene of type I collagen produces a lethal form of osteogenesis imperfecta. J. Biol. Chem. 1984, 259, 11129–11138. [Google Scholar] [CrossRef]
- Bonadio, J.; Byers, P.H. Subtle structural alterations in the chains of type I procollagen produce osteogenesis imperfecta type II. Nature 1985, 316, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Byers, P.H.; Tsipouras, P.; Bonadio, J.F.; Starman, B.J.; Schwartz, R.C. Perinatal lethal osteogenesis imperfecta (OI type II): A biochemically heterogeneous disorder usually due to new mutations in the genes for type I collagen. Am. J. Hum. Genet. 1988, 42, 237–248. [Google Scholar] [PubMed]
- Nicholls, A.C.; Oliver, J.; Renouf, D.V.; Keston, M.; Pope, F.M. Substitution of cysteine for glycine at residue 415 of one allele of the alpha 1(I) chain of type I procollagen in type III/IV osteogenesis imperfecta. J. Med. Genet. 1991, 28, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.; Narcisi, P.; Lloyd, J.; Ferguson, C.; Pope, F.M. The substitution of glycine 661 by arginine in type III collagen produces mutant molecules with different thermal stabilities and causes Ehlers-Danlos syndrome type IV. J. Med. Genet. 1993, 30, 690–693. [Google Scholar] [CrossRef] [PubMed]
- Marini, J.C.; Forlino, A.; Cabral, W.A.; Barnes, A.M.; San Antonio, J.D.; Milgrom, S.; Hyland, J.C.; Körkkö, J.; Prockop, D.J.; De Paepe, A.; et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: Regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 2007, 28, 209–221. [Google Scholar] [CrossRef]
- Dombrowski, K.E.; Vogel, B.E.; Prockop, D.J. Mutations that alter the primary structure of type I procollagen have long-range effects on its cleavage by procollagen N-proteinase. Biochemistry 1989, 28, 7107–7112. [Google Scholar] [CrossRef]
- Makareeva, E.; Cabral, W.A.; Marini, J.C.; Leikin, S. Molecular mechanism of alpha 1(I)-osteogenesis imperfecta/Ehlers-Danlos syndrome: Unfolding of an N-anchor domain at the N-terminal end of the type I collagen triple helix. J. Biol. Chem. 2006, 281, 6463–6470. [Google Scholar] [CrossRef]
- Bateman, J.F.; Shoulders, M.D.; Lamandé, S.R. Collagen misfolding mutations: The contribution of the unfolded protein response to the molecular pathology. Connect. Tissue Res. 2022, 63, 210–227. [Google Scholar] [CrossRef]
- Richards, A.J.; Yates, J.R.; Williams, R.; Payne, S.J.; Pope, F.M.; Scott, J.D.; Snead, M.P. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum. Mol. Genet. 1996, 5, 1339–1343. [Google Scholar] [CrossRef]
- Richards, A.J.; Martin, S.; Nicholls, A.C.; Harrison, J.B.; Pope, F.M.; Burrows, N.P. A single base mutation in COL5A2 causes Ehlers-Danlos syndrome type II. J. Med. Genet. 1998, 35, 846–848. [Google Scholar] [CrossRef]
- Pihlajamaa, T.; Prockop, D.J.; Faber, J.; Winterpacht, A.; Zabel, B.; Giedion, A.; Wiesbauer, P.; Spranger, J.; Ala-Kokko, L. Heterozygous glycine substitution in the COL11A2 gene in the original patient with the Weissenbacher-Zweymüller syndrome demonstrates its identity with heterozygous OSMED (nonocular Stickler syndrome). Am. J. Med. Genet. 1998, 80, 115–120. [Google Scholar] [CrossRef]
- Ritelli, M.; Dordoni, C.; Venturini, M.; Chiarelli, N.; Quinzani, S.; Traversa, M.; Zoppi, N.; Vascellaro, A.; Wischmeijer, A.; Manfredini, E.; et al. Clinical and molecular characterization of 40 patients with classic Ehlers-Danlos syndrome: Identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J. Rare Dis. 2013, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Tompson, S.W.; Bacino, C.A.; Safina, N.P.; Bober, M.B.; Proud, V.K.; Funari, T.; Wangler, M.F.; Nevarez, L.; Ala-Kokko, L.; Wilcox, W.R.; et al. Fibrochondrogenesis results from mutations in the COL11A1 type XI collagen gene. Am. J. Hum. Genet. 2010, 87, 708–712. [Google Scholar] [CrossRef]
- Vikkula, M.; Mariman, E.C.; Lui, V.C.; Zhidkova, N.I.; Tiller, G.E.; Goldring, M.B.; van Beersum, S.E.; de Waal Malefijt, M.C.; van den Hoogen, F.H.; Ropers, H.H. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell 1995, 80, 431–437. [Google Scholar] [CrossRef]
- Mayne, R.; Brewton, R.G.; Mayne, P.M.; Baker, J.R. Isolation and characterization of the chains of type V/type XI collagen present in bovine vitreous. J. Biol. Chem. 1993, 268, 9381–9386. [Google Scholar] [CrossRef]
- Ellard, S.; Baple, E.L.; Callaway, A.; Berry, I.; Forrester, N.; Turnbull, C.; Owens, M.; Eccles, D.M.; Abbs, S.; Scott, R.; et al. ACGS Best Practice Guidelines for Variant Classification. 2020. Available online: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (accessed on 23 May 2022).
- Williams, C.J.; Considine, E.L.; Knowlton, R.G.; Reginato, A.; Neumann, G.; Harrison, D.; Buxton, P.; Jimenez, S.; Prockop, D.J. Spondyloepiphyseal dysplasia and precocious osteoarthritis in a family with an Arg75-->Cys mutation in the procollagen type II gene (COL2A1). Hum. Genet. 1993, 92, 499–505. [Google Scholar] [CrossRef]
- Hoornaert, K.P.; Marik, I.; Kozlowski, K.; Cole, T.; Le Merrer, M.; Leroy, J.G.; Coucke, P.J.; Sillence, D.; Mortier, G.R. Czech dysplasia metatarsal type: Another type II collagen disorder. Eur. J. Hum. Genet. 2007, 15, 1269–1275. [Google Scholar] [CrossRef]
- Richards, A.J.; Baguley, D.M.; Yates, J.R.; Lane, C.; Nicol, M.; Harper, P.S.; Scott, J.D.; Snead, M.P. Variation in the vitreous phenotype of Stickler syndrome can be caused by different amino acid substitutions in the X position of the type II collagen Gly-X-Y triple helix. Am. J. Hum. Genet. 2000, 67, 1083–1094. [Google Scholar] [CrossRef][Green Version]
- Sulko, J.; Czarny-Ratajczak, M.; Wozniak, A.; Latos-Bielenska, A.; Kozlowski, K. Novel amino acid substitution in the Y-position of collagen type II causes spondyloepimetaphyseal dysplasia congenita. Am. J. Med. Genet. A 2005, 137A, 292–297. [Google Scholar] [CrossRef]
- Chen, L.; Yang, W.; Cole, W.G. Alternative splicing of exon 12 of the COL2A1 gene interrupts the triple helix of type-II collagen in the Kniest form of spondyloepiphyseal dysplasia. J. Orthop. Res. 1996, 14, 712–721. [Google Scholar] [CrossRef]
- Khajavi, M.; Inoue, K.; Lupski, J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006, 14, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Kuivaniemi, H.; Tromp, G.; Prockop, D.J. Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels. Hum. Mutat. 1997, 9, 300–315. [Google Scholar] [CrossRef]
- Hoornaert, K.P.; Vereecke, I.; Dewinter, C.; Rosenberg, T.; Beemer, F.A.; Leroy, J.G.; Bendix, L.; Björck, E.; Bonduelle, M.; Boute, O.; et al. Stickler syndrome caused by COL2A1 mutations: Genotype-phenotype correlation in a series of 100 patients. Eur. J. Hum. Genet. 2010, 18, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Zankl, A.; Zabel, B.; Hilbert, K.; Wildhardt, G.; Cuenot, S.; Xavier, B.; Ha-Vinh, R.; Bonafé, L.; Spranger, J.; Superti-Furga, A. Spondyloperipheral dysplasia is caused by truncating mutations in the C-propeptide of COL2A1. Am. J. Med. Genet. A 2004, 129A, 144–148. [Google Scholar] [CrossRef]
- Leistritz, D.F.; Pepin, M.G.; Schwarze, U.; Byers, P.H. COL3A1 haploinsufficiency results in a variety of Ehlers-Danlos syndrome type IV with delayed onset of complications and longer life expectancy. Genet. Med. 2011, 13, 717–722. [Google Scholar] [CrossRef]
- Schwarze, U.; Atkinson, M.; Hoffman, G.G.; Greenspan, D.S.; Byers, P.H. Null alleles of the COL5A1 gene of type V collagen are a cause of the classical forms of Ehlers-Danlos syndrome (types I and II). Am. J. Hum. Genet. 2000, 66, 1757–1765. [Google Scholar] [CrossRef][Green Version]
- Melkoniemi, M.; Brunner, H.G.; Manouvrier, S.; Hennekam, R.; Superti-Furga, A.; Kääriäinen, H.; Pauli, R.M.; van Essen, T.; Warman, M.L.; Bonaventure, J.; et al. Autosomal recessive disorder otospondylomegaepiphyseal dysplasia is associated with loss-of-function mutations in the COL11A2 gene. Am. J. Hum. Genet. 2000, 66, 368–377. [Google Scholar] [CrossRef]
- Richards, A.J.; Laidlaw, M.; Meredith, S.P.; Shankar, P.; Poulson, A.V.; Scott, J.D.; Snead, M.P. Missense and silent mutations in COL2A1 result in Stickler syndrome but via different molecular mechanisms. Hum. Mutat. 2007, 28, 639. [Google Scholar] [CrossRef]
- Richards, A.J.; McNinch, A.; Whittaker, J.; Treacy, B.; Oakhill, K.; Poulson, A.; Snead, M.P. Splicing analysis of unclassified variants in COL2A1 and COL11A1 identifies deep intronic pathogenic mutations. Eur. J. Hum. Genet. 2012, 20, 552–558. [Google Scholar] [CrossRef][Green Version]
- Sandell, L.J.; Morris, N.; Robbins, J.R.; Goldring, M.B. Alternatively spliced type II procollagen mRNAs define distinct populations of cells during vertebral development: Differential expression of the amino-propeptide. J. Cell Biol. 1991, 114, 1307–1319. [Google Scholar] [CrossRef]
- Zhidkova, N.I.; Justice, S.K.; Mayne, R. Alternative mRNA processing occurs in the variable region of the pro-alpha 1(XI) and pro-alpha 2(XI) collagen chains. J. Biol. Chem. 1995, 270, 9486–9493. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.J.; Martin, S.; Yates, J.R.; Scott, J.D.; Baguley, D.M.; Pope, F.M.; & Snead, M.P. COL2A1 exon 2 mutations: Relevance to the Stickler and Wagner syndromes. Br. J. Ophthalmol. 2000, 84, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.J.; Fincham, G.S.; McNinch, A.; Hill, D.; Poulson, A.V.; Castle, B.; Lees, M.M.; Moore, A.T.; Scott, J.D.; Snead, M.P. Alternative splicing modifies the effect of mutations in COL11A1 and results in recessive type 2 Stickler syndrome with profound hearing loss. J. Med. Genet. 2013, 50, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Richards, A.J.; Yates, J.R.; Scott, J.D.; Pope, M.; Snead, M.P. Stickler syndrome: Further mutations in COL11A1 and evidence for additional locus heterogeneity. Eur. J. Hum. Genet. 1999, 7, 807–814. [Google Scholar] [CrossRef]
- Lees, J.F.; Bulleid, N.J. The role of cysteine residues in the folding and association of the COOH-terminal propeptide of types I and III procollagen. J. Biol. Chem. 1994, 269, 24354–24360. [Google Scholar] [CrossRef]
- Lindahl, K.; Barnes, A.M.; Fratzl-Zelman, N.; Whyte, M.P.; Hefferan, T.E.; Makareeva, E.; Brusel, M.; Yaszemski, M.J.; Rubin, C.J.; Kindmark, A.; et al. COL1 C-propeptide cleavage site mutations cause high bone mass osteogenesis imperfecta. Hum. Mutat. 2011, 32, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, J.M.; Mariano, N.; Zhao, Y.; Harlos, K.; Exposito, J.Y.; Jones, E.Y.; Moali, C.; Aghajari, N.; Hulmes, D.J. Structural basis of fibrillar collagen trimerization and related genetic disorders. Nat. Struct. Mol. Biol. 2012, 19, 1031–1036. [Google Scholar] [CrossRef]
- Stembridge, N.S.; Vandersteen, A.M.; Ghali, N.; Sawle, P.; Nesbitt, M.; Pollitt, R.C.; Ferguson, D.J.; Holden, S.; Elmslie, F.; Henderson, A.; et al. Clinical, structural, biochemical and X-ray crystallographic correlates of pathogenicity for variants in the C-propeptide region of the COL3A1 gene. Am. J. Med. Genet. A 2015, 167A, 1763–1772. [Google Scholar] [CrossRef]
- Byers, P.H.; Duvic, M.; Atkinson, M.; Robinow, M.; Smith, L.T.; Krane, S.M.; Greally, M.T.; Ludman, M.; Matalon, R.; Pauker, S.; et al. Ehlers-Danlos syndrome type VIIA and VIIB result from splice-junction mutations or genomic deletions that involve exon 6 in the COL1A1 and COL1A2 genes of type I collagen. Am. J. Med. Genet. 1997, 72, 94–105. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Schwarze, U.; Jennings, J.F.; Byers, P.H. Molecular mechanisms of classical Ehlers-Danlos syndrome (EDS). Hum. Mutat. 2009, 30, 995–1002. [Google Scholar] [CrossRef][Green Version]
- Cho, S.Y.; Lee, J.H.; Ki, C.S.; Chang, M.S.; Jin, D.K.; Han, H.S. Osteogenesis imperfecta Type I caused by a novel mutation in the start codon of the COL1A1 gene in a Korean family. Ann. Clin. Lab. Sci. 2015, 45, 100–105. [Google Scholar] [PubMed]
- Lindert, U.; Gnoli, M.; Maioli, M.; Bedeschi, M.F.; Sangiorgi, L.; Rohrbach, M.; Giunta, C. Insight into the Pathology of a COL1A1 Signal Peptide Heterozygous Mutation Leading to Severe Osteogenesis Imperfecta. Calcif. Tissue Int. 2018, 102, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Symoens, S.; Malfait, F.; Renard, M.; André, J.; Hausser, I.; Loeys, B.; Coucke, P.; De Paepe, A. COL5A1 signal peptide mutations interfere with protein secretion and cause classic Ehlers-Danlos syndrome. Hum. Mutat. 2009, 30, E395–E403. [Google Scholar] [CrossRef] [PubMed]
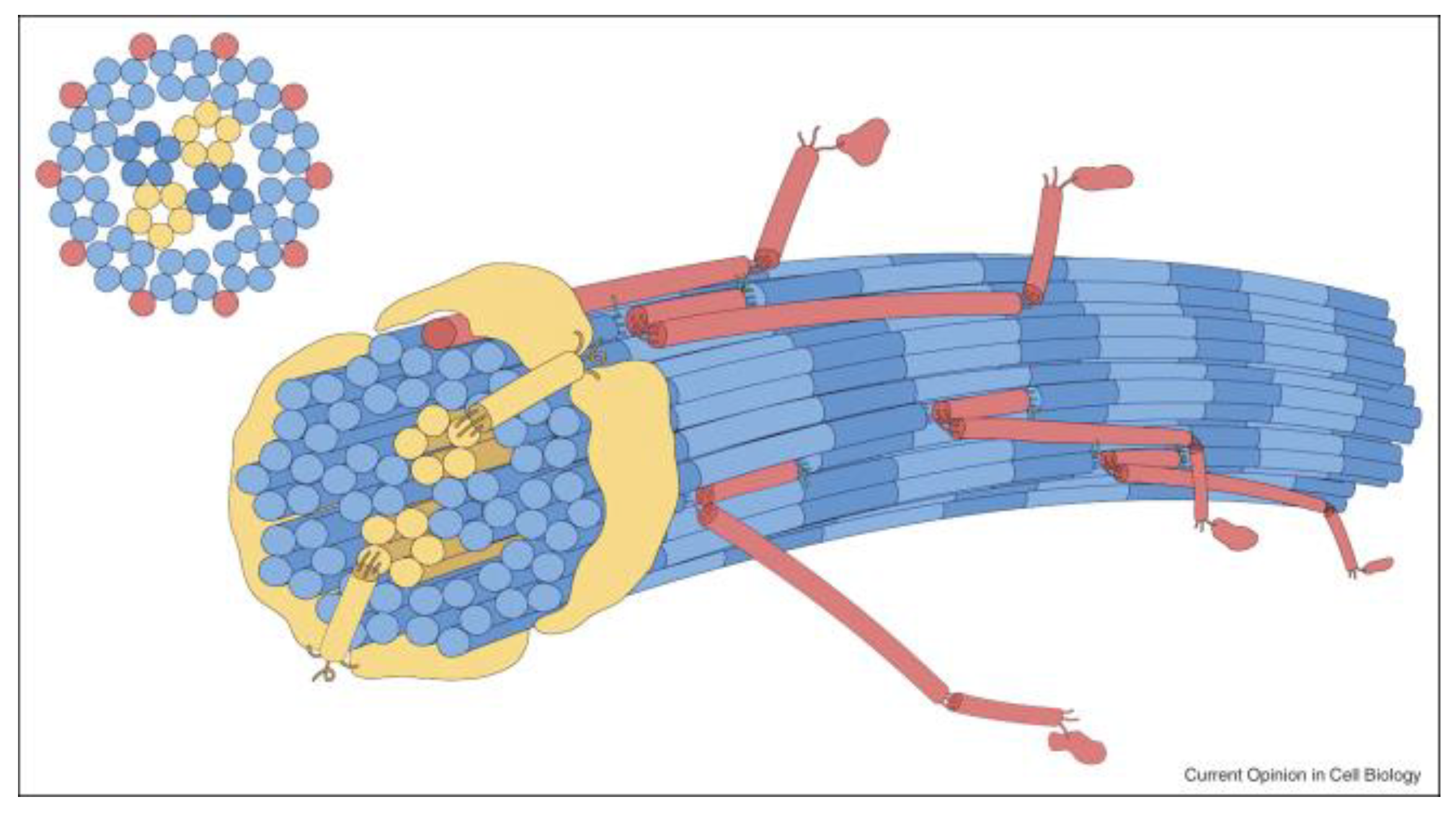
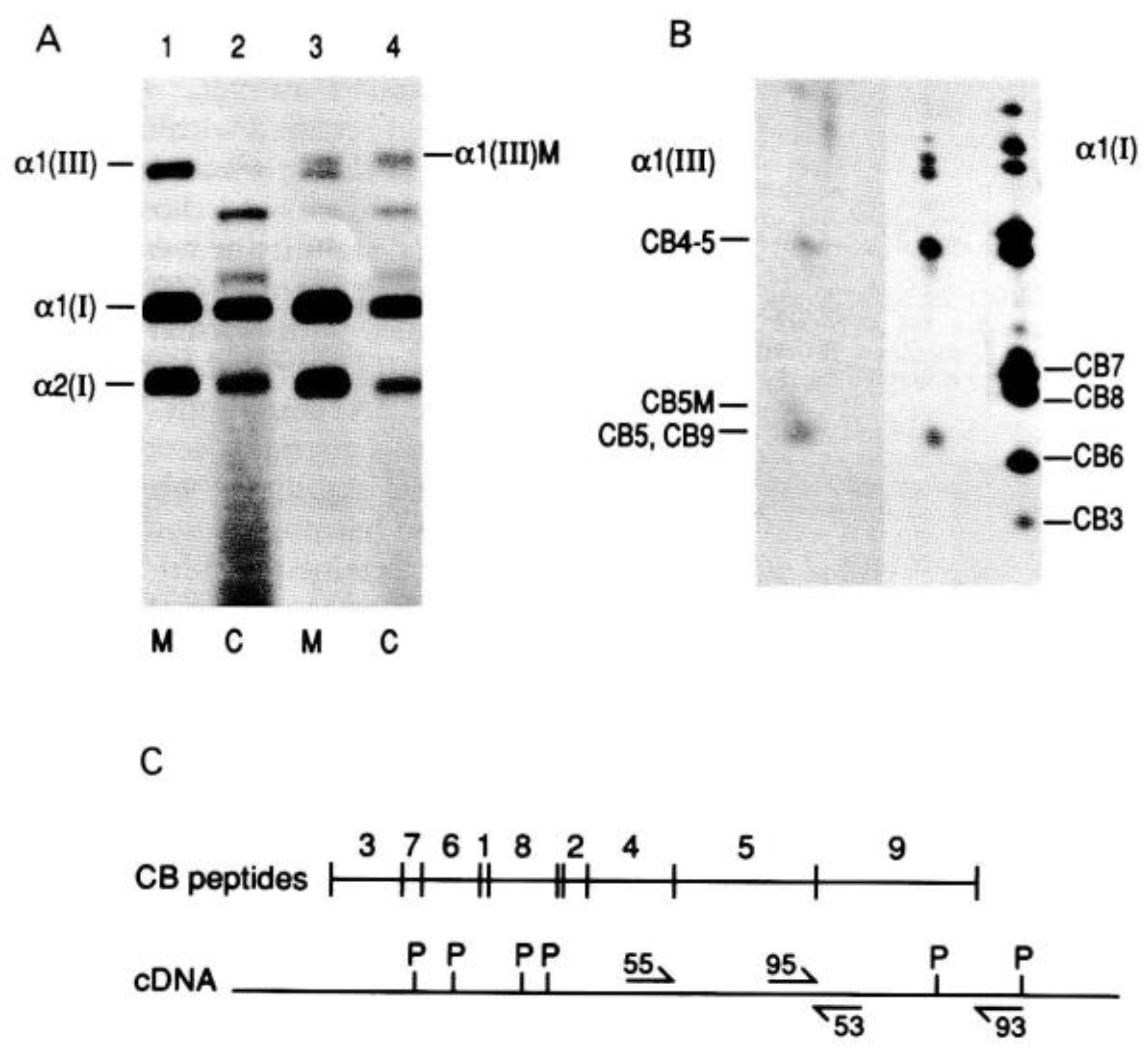
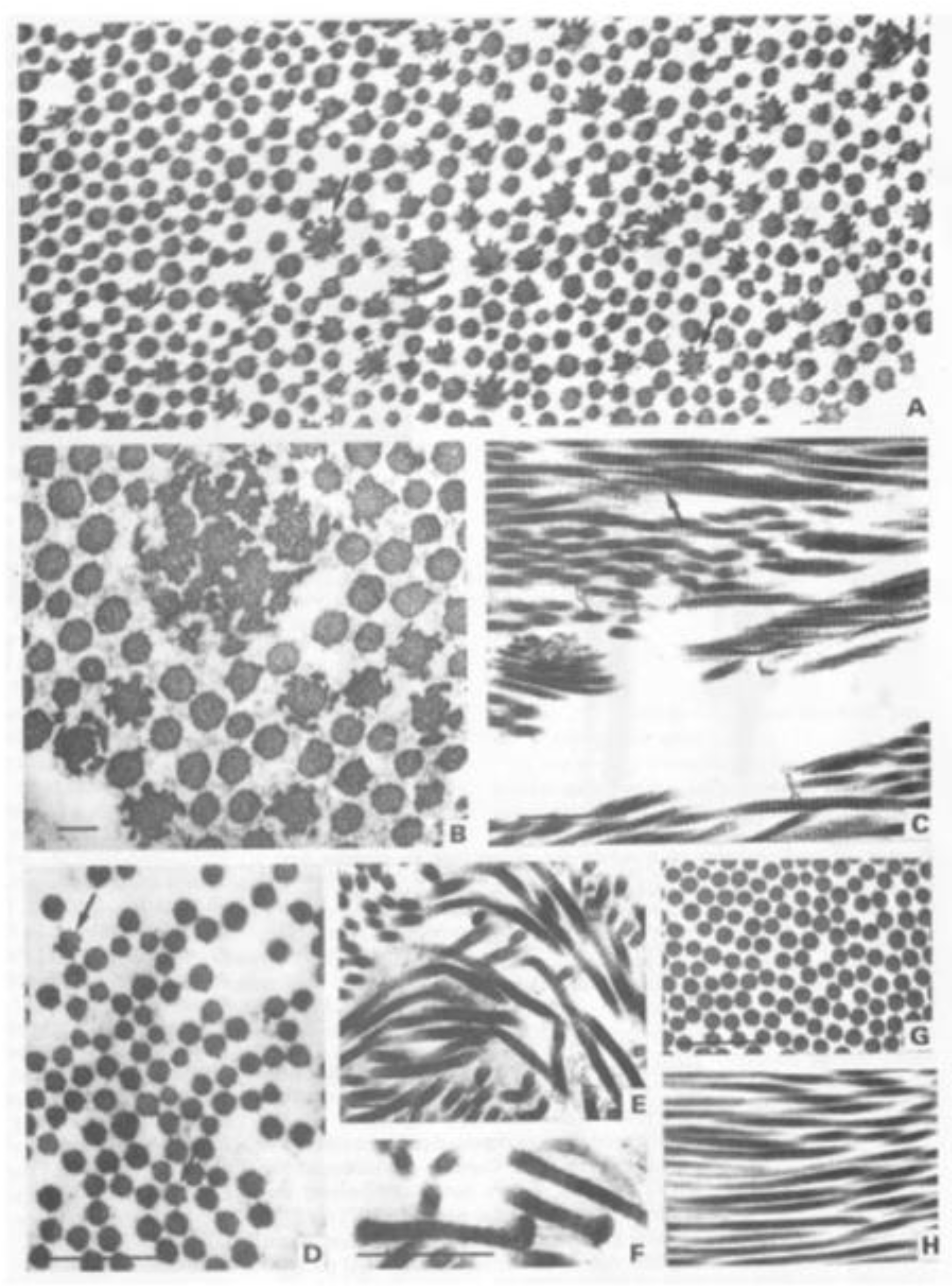
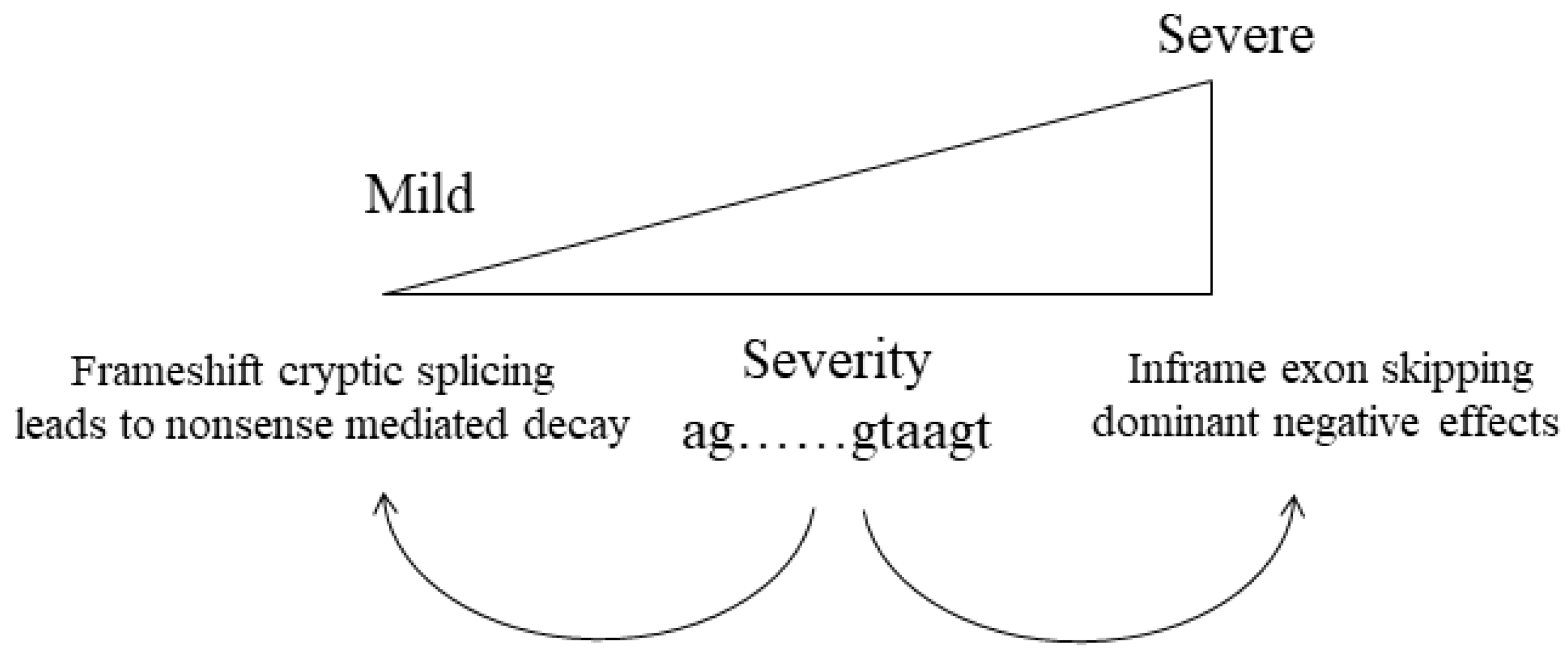
| Collagen Type | Stoichiometry | Tissue | Genes |
|---|---|---|---|
| Type I | α1(I)2α2(I) | Multiple tissues including Bone, Skin, Tendon | COL1A1, COL1A2 |
| Type II | α1(II)3 | Cartilage, Vitreous | COL2A1 |
| Type III | α1(III)3 | Multiple tissues including Skin, Blood vessels | COL3A1 |
| Type V | Major form α1(V)2α2(V) Placenta α1(V)α2(V)α3(V) | Co-expressed with Types I and III collagens | COL5A1 COL5A2 COL5A3 |
| Type XI | Cartilage α1(XI)α2(XI)α3(XI) Vitreous α1(XI)2α3(XI) ?? α1(XI)2α2(V) ?? | Co-expressed with Type II collagen | COL11A1 COL11A2 COL2A1 COL5A2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richards, A.J.; Snead, M.P. Molecular Basis of Pathogenic Variants in the Fibrillar Collagens. Genes 2022, 13, 1199. https://doi.org/10.3390/genes13071199
Richards AJ, Snead MP. Molecular Basis of Pathogenic Variants in the Fibrillar Collagens. Genes. 2022; 13(7):1199. https://doi.org/10.3390/genes13071199
Chicago/Turabian StyleRichards, Allan J., and Martin P. Snead. 2022. "Molecular Basis of Pathogenic Variants in the Fibrillar Collagens" Genes 13, no. 7: 1199. https://doi.org/10.3390/genes13071199
APA StyleRichards, A. J., & Snead, M. P. (2022). Molecular Basis of Pathogenic Variants in the Fibrillar Collagens. Genes, 13(7), 1199. https://doi.org/10.3390/genes13071199






