Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Clinical Characteristics of the Study Subjects
2.3. Sample Collection
2.4. Exosome Isolation from Human Serum
2.5. Serum and Exosomal miRNA Next-Generation Sequencing (NGS)
2.6. Differential miRNA Expression Analysis
2.7. Statistical Analysis
3. Results
3.1. Profile of Circulating Serum miRNA Expression in SGA-nCU and AGA
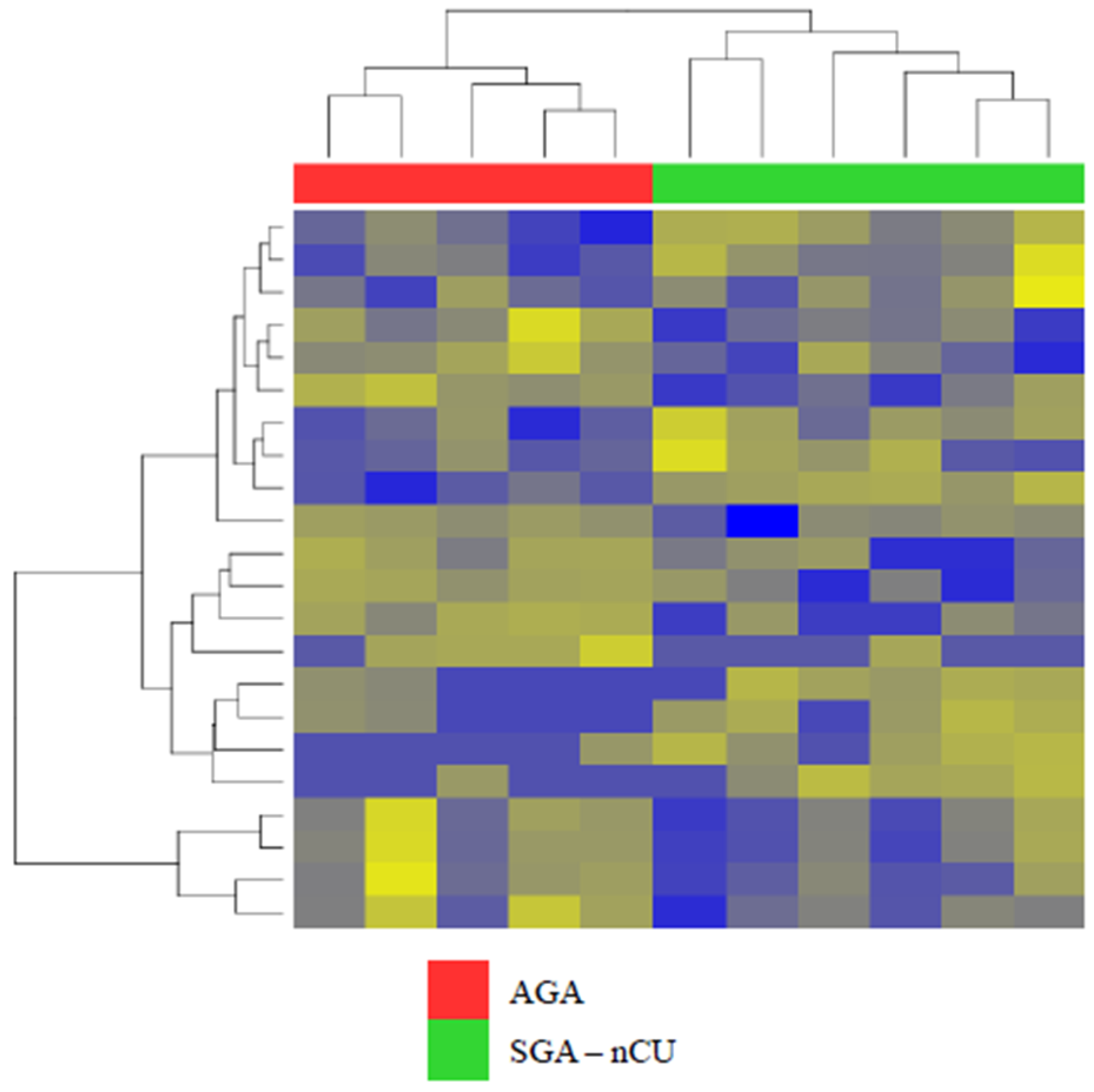
3.2. Exosomal miRNA Profiles Show Different Expression Patterns than Those Collected from Serum
3.3. Exosomal miRNA Profiles of SGA-CU Were More Similar to Those of AGA than Those of SGA-nCU
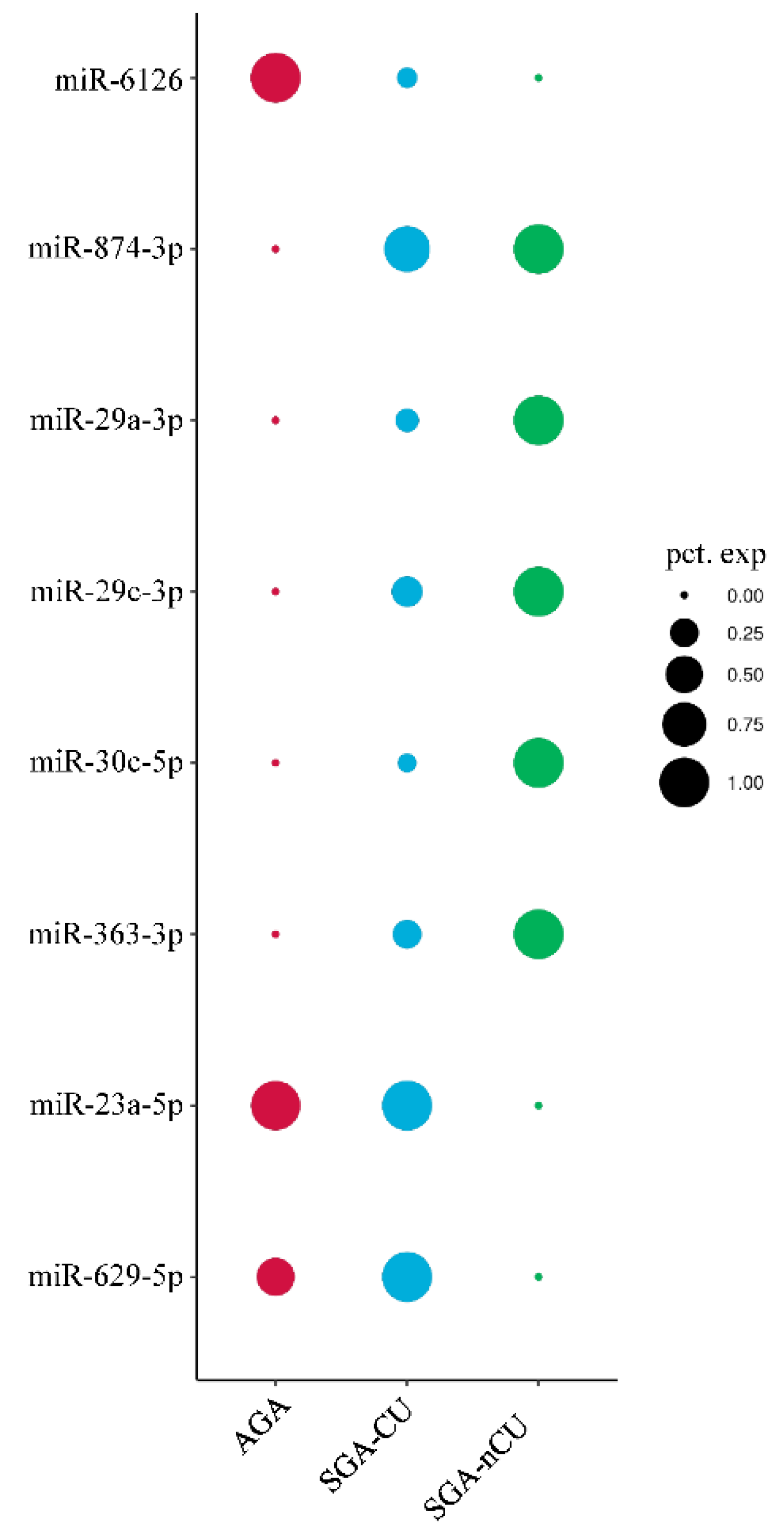
3.4. Exosomal miRNA Profiles of SGA-nCU and SGA-CU Show Potential Differences
3.5. Comparison of the Exosomal miRNA Profiles of AGA, SGA-CU, and SGA-nCU Children
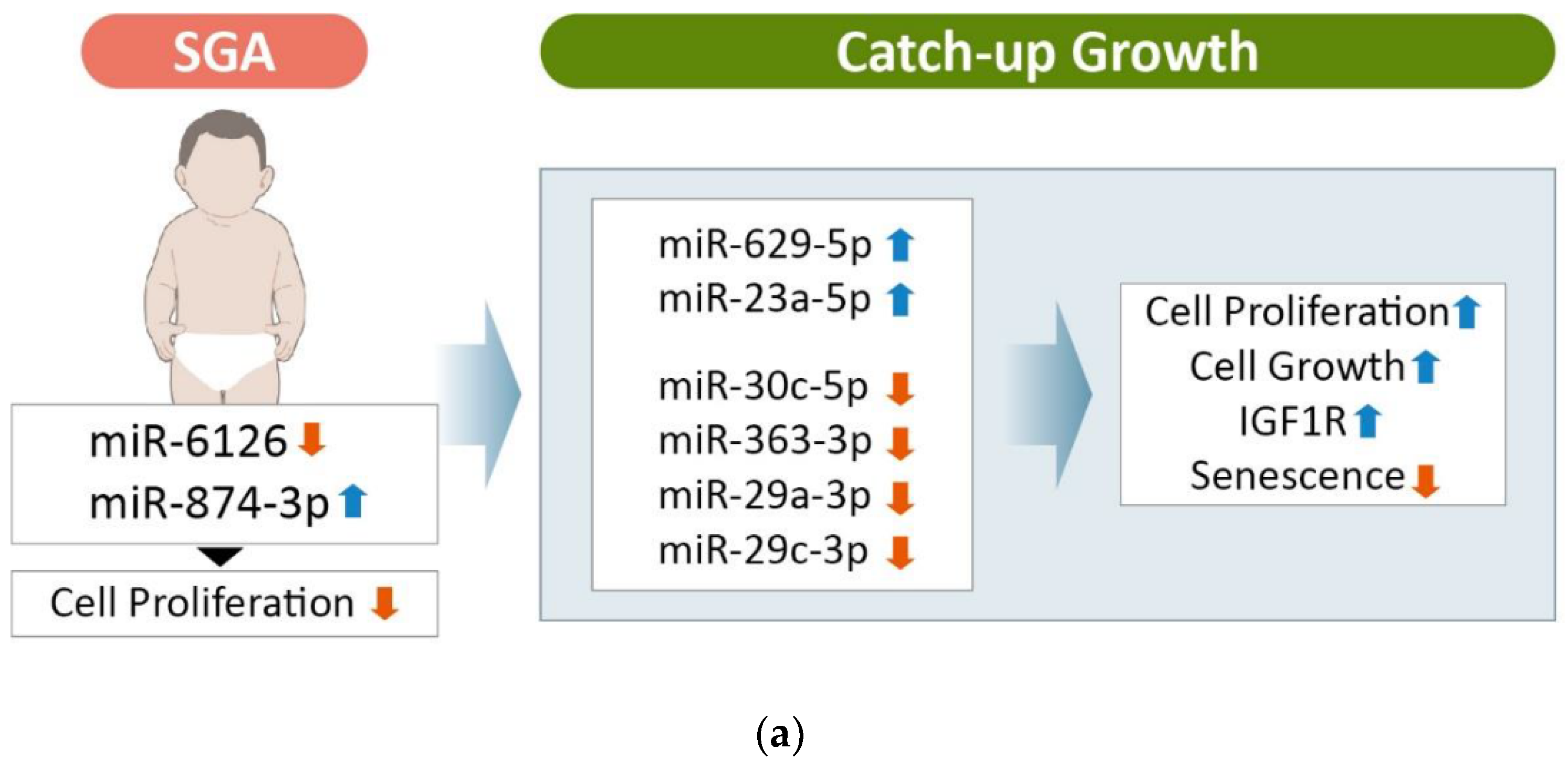
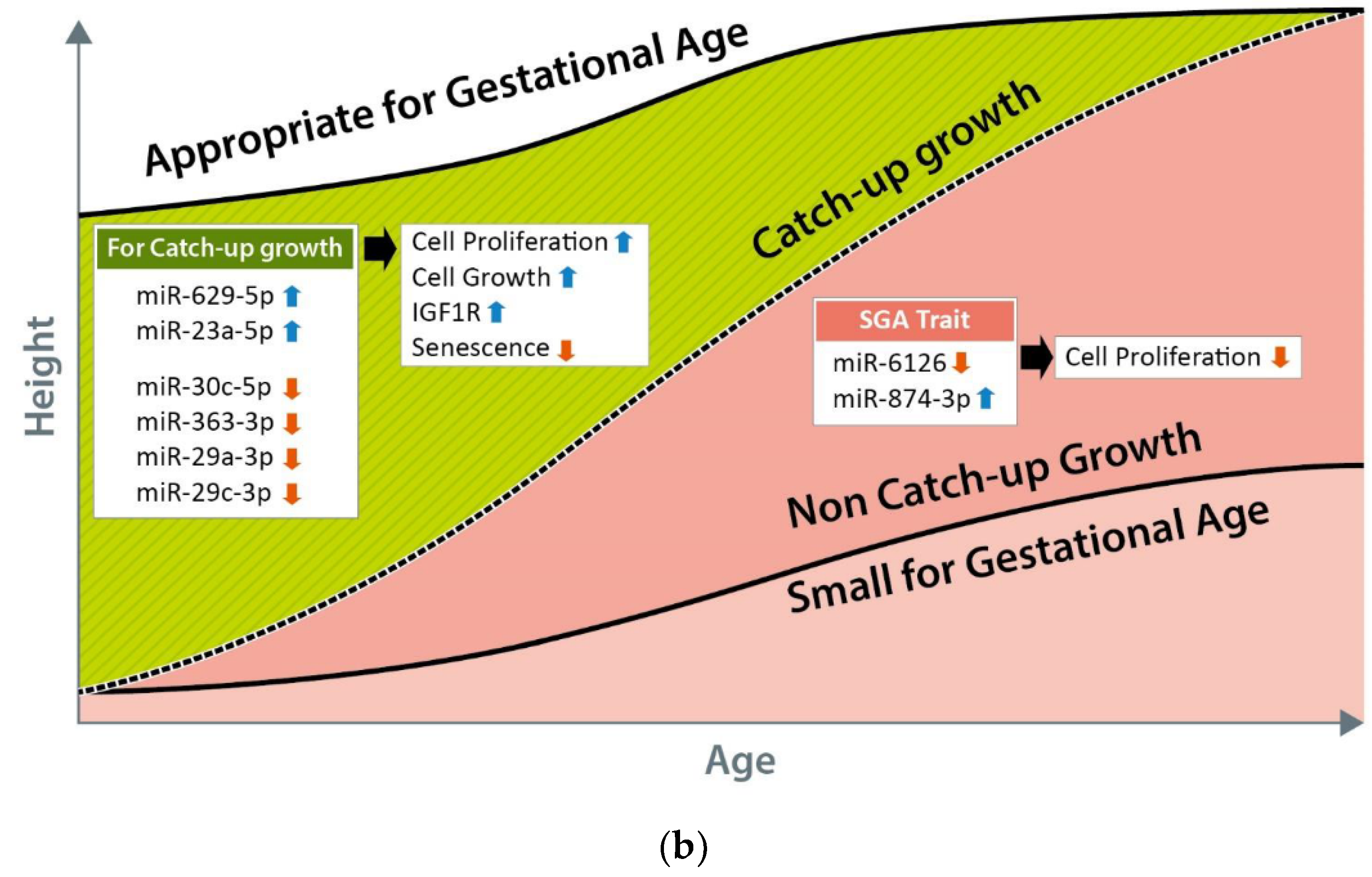
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Clayton, P.; Cianfarani, S.; Czernichow, P.; Johannsson, G.; Rapaport, R.; Rogol, A. Management of the Child Born Small for Gestational Age through to Adulthood: A Consensus Statement of the International Societies of Pediatric Endocrinology and the Growth Hormone Research Society. J. Clin. Endocrinol. Metab. 2007, 92, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, F.C.; Lubchenco, L.O. A practical classification of newborn infants by weight and gestational age. J. Pediatr. 1967, 71, 159–163. [Google Scholar] [CrossRef]
- Finken, M.J.J.; Van Der Steen, M.; Smeets, C.C.J.; Walenkamp, M.J.E.; De Bruin, C.; Hokken-Koelega, A.C.S.; Wit, J.M. Children Born Small for Gestational Age: Differential Diagnosis, Molecular Genetic Evaluation, and Implications. Endocr. Rev. 2018, 39, 851–894. [Google Scholar] [CrossRef] [PubMed]
- Campisi, S.C.; Carbone, S.; Zlotkin, S. Catch-Up Growth in Full-Term Small for Gestational Age Infants: A Systematic Review. Adv. Nutr. Int. Rev. J. 2019, 10, 104–111. [Google Scholar] [CrossRef]
- Stalman, S.E.; Solanky, N.; Ishida, M.; Alemán-Charlet, C.; Abu-Amero, S.; Alders, M.; Alvizi, L.; Baird, W.; Demetriou, C.; Henneman, P.; et al. Genetic Analyses in Small-for-Gestational-Age Newborns. J. Clin. Endocrinol. Metab. 2018, 103, 917–925. [Google Scholar] [CrossRef]
- Lee, A.C.; Kozuki, N.; Cousens, S.; Stevens, G.A.; Blencowe, H.; Silveira, M.F.; Sania, A.; Rosen, H.E.; Schmiegelow, C.; Adair, L.S.; et al. Estimates of burden and consequences of infants born small for gestational age in low and middle income countries with INTERGROWTH-21(st) standard: Analysis of CHERG datasets. BMJ 2017, 358, j3677. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, D.H.; Lim, J.S. Growth status of children and adolescents born small for gestational age at full term in Korea: Data from the KNHANES-V. J. Pediatr. Endocrinol. Metab. 2020, 33, 743–750. [Google Scholar] [CrossRef]
- Van Wassenaer, A. Neurodevelopmental consequences of being born SGA. Pediatr. Endocrinol. Rev. 2005, 2, 372–377. [Google Scholar]
- Milovanovic, I.; Njuieyon, F.; Deghmoun, S.; Chevenne, D.; Levy-Marchal, C.; Beltrand, J. SGA Children with Moderate Catch-Up Growth Are Showing the Impaired Insulin Secretion at the Age of 4. PLoS ONE 2014, 9, e100337. [Google Scholar] [CrossRef]
- Hong, Y.H.; Chung, S. Small for gestational age and obesity related comorbidities. Ann. Pediatr. Endocrinol. Metab. 2018, 23, 4–8. [Google Scholar] [CrossRef]
- Verkauskiene, R.; Petraitiene, I.; Wikland, K.A. Puberty in Children Born Small for Gestational Age. Horm. Res. Paediatr. 2013, 80, 69–77. [Google Scholar] [CrossRef]
- Lee, P.A.; Chernausek, S.D.; Hokken-Koelega, A.C.S.; Czernichow, P.; The International SGA Advisory Board. International Small for Gestational Age Advisory Board Consensus Development Conference Statement: Management of Short Children Born Small for Gestational Age, April 24–October 1, 2001. Pediatrics 2003, 111, 1253–1261. [Google Scholar] [CrossRef]
- Karlberg, J.; Albertsson-Wikland, K.; Kwan, E.; Lam, B.; Low, L. The Timing of Early Postnatal Catch-Up Growth in Normal, Full-Term Infants Born Short for Gestational Age. Horm. Res. 1997, 48, 17–24. [Google Scholar] [CrossRef]
- Paz, I.; Seidman, D.S.; Danon, Y.L.; Laor, A.; Stevenson, D.K.; Gale, R. Are children born small for gestational age at increased risk of short stature? Am. J. Dis. Child. 1993, 147, 337–339. [Google Scholar] [CrossRef]
- Karlberg, J.; Albertsson-Wikland, K. Growth in Full-Term Small-for-Gestational-Age Infants: From Birth to Final Height. Pediatr. Res. 1995, 38, 733–739. [Google Scholar] [CrossRef]
- Van Dijk, M.; Mulder, P.; Houdijk, M.; Mulder, J.; Noordam, K.; Odink, R.J.; Rongen-Westerlaken, C.; Voorhoeve, P.; Waelkens, J.; Stokvis-Brantsma, J.; et al. High serum levels of growth hormone (GH) and insulin-like growth factor-I (IGF-I) during high-dose GH treatment in short children born small for gestational age. J. Clin. Endocrinol. Metab. 2006, 91, 1390–1396. [Google Scholar] [CrossRef][Green Version]
- Iñiguez, G.; Ong, K.; Bazaes, R.; Avila, A.; Salazar, T.; Dunger, D.; Mericq, V. Longitudinal Changes in Insulin-Like Growth Factor-I, Insulin Sensitivity, and Secretion from Birth to Age Three Years in Small-for-Gestational-Age Children. J. Clin. Endocrinol. Metab. 2006, 91, 4645–4649. [Google Scholar] [CrossRef]
- Gat-Yablonski, G.; Phillip, M. Nutritionally-Induced Catch-Up Growth. Nutrients 2015, 7, 517–551. [Google Scholar] [CrossRef]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.T.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA Profiles in Patients with Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas-García, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef]
- Galasso, M.; Sana, M.E.; Volinia, S. Non-coding RNAs: A key to future personalized molecular therapy? Genome Med. 2010, 2, 12. [Google Scholar] [CrossRef]
- Mas-Parés, B.; Xargay-Torrent, S.; Bonmatí, A.; Lizarraga-Mollinedo, E.; Martínez-Calcerrada, J.M.; Carreras-Badosa, G.; Prats-Puig, A.; de Zegher, F.; Ibáñez, L.; López-Bermejo, A.; et al. Umbilical cord microRNA in small-for-gestational age children and association with catch-up growth: A pilot study. J. Clin. Endocrinol. Metab. 2019, 104, 5285–5298. [Google Scholar] [CrossRef]
- Marzano, F.; Faienza, M.F.; Caratozzolo, M.F.; Brunetti, G.; Chiara, M.; Horner, D.S.; Annese, A.; D’Erchia, A.M.; Consiglio, A.; Pesole, G.; et al. Pilot study on circulating miRNA signature in children with obesity born small for gestational age and appropriate for gestational age. Pediatr. Obes. 2018, 13, 803–811. [Google Scholar] [CrossRef]
- Lim, J.S.; Lim, S.W.; Ahn, J.H.; Song, B.S.; Shim, K.S.; Hwang, I.T. New Korean reference for birth weight by gestational age and sex: Data from the Korean Statistical Information Service (2008–2012). Ann. Pediatr. Endocrinol. Metab. 2014, 19, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Inzaghi, E.; Kistner, A.; Germani, D.; Deodati, A.; Vanpee, M.; Legnevall, L.; Berinder, K.; Cianfarani, S. A prospective case-control study on miRNA circulating levels in subjects born small for gestational age (SGA) evaluated from childhood into young adulthood. PLoS ONE 2020, 15, e0228075. [Google Scholar] [CrossRef]
- Onyshchenko, K.V.; Voitsitskyi, T.V.; Grygorenko, V.M.; Saidakova, N.O.; Pereta, L.V.; Onyschuk, A.P.; Skrypkina, I.Y. Expression of micro-RNA hsa-miR-30c-5p and hsa-miR-138-1 in renal cell carcinoma. Exp. Oncol. 2020, 42, 115–119. [Google Scholar]
- Song, S.; Long, M.; Yu, G.; Cheng, Y.; Yang, Q.; Liu, J.; Wang, Y.; Sheng, J.; Wang, L.; Wang, Z.; et al. Urinary exosome miR-30c-5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5. J. Cell. Mol. Med. 2019, 23, 6755–6765. [Google Scholar] [CrossRef]
- Gao, B.-H.; Wu, H.; Wang, X.; Ji, L.-L.; Chen, C. MiR-30c-5p inhibits high glucose-induced EMT and renal fibrogenesis by down-regulation of JAK1 in diabetic nephropathy. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1338–1349. [Google Scholar]
- Gao, C.; Qian, H.; Shi, Q.; Zhang, H. MicroRNA-363-3p serves as a diagnostic biomarker of acute myocardial infarction and regulates vascular endothelial injury by targeting KLF2. Cardiovasc. Diagn. Ther. 2020, 10, 421–430. [Google Scholar] [CrossRef]
- Zheng, J.; Zhu, S.; Xu, H.; Li, J.; Tang, H.; Zhou, Y.; Huang, Z.; Liu, G. miR-363-3p inhibits rat lung alveolar type II cell proliferation by downregulating STRA6 expression and induces cell apoptosis via cellular oxidative stress and G1-phase cell cycle arrest. Transl. Pediatr. 2021, 10, 2095–2105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, T.; Huang, H.; Jiang, Y.; Yang, L.; Lin, Z.; He, H.; Liu, T.; Wu, B.; Chen, J.; et al. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget 2017, 8, 20133–20144. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, J.; Hu, H.; Duan, Q.; Chen, J.; Li, L. MiR-363-3p attenuates neonatal hypoxic-ischemia encephalopathy by targeting DUSP5. Neurosci. Res. 2021, 171, 103–113. [Google Scholar] [CrossRef]
- Dong, J.; Geng, J.; Tan, W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed. Pharmacother. 2018, 105, 922–931. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Cao, L.; Zhang, T.; Yue, D.; Wang, L.; Ping, Y.; He, Q.; Zhang, C.; Wang, M.; et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget 2017, 8, 86592–86603. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, W.; Wu, Z.; Liu, Y.; Shi, Y.; Gong, J.; Shen, W.; Liu, C. miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- Zhang, H.-W.; Wang, E.-W.; Li, L.-X.; Yi, S.-H.; Li, L.-C.; Xu, F.-L.; Wang, D.-L.; Wu, Y.-Z.; Nian, W.-Q. A regulatory loop involving miR-29c and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget 2016, 7, 85905–85916. [Google Scholar] [CrossRef]
- Niu, M.; Gao, D.; Wen, Q.; Wei, P.; Pan, S.; Shuai, C.; Ma, H.; Xiang, J.; Li, Z.; Fan, S.; et al. MiR-29c regulates the expression of miR-34c and miR-449a by targeting DNA methyltransferase 3a and 3b in nasopharyngeal carcinoma. BMC Cancer 2016, 16, 218. [Google Scholar] [CrossRef]
- Shang, J.; Yao, Y.; Fan, X.; Shangguan, L.; Li, J.; Liu, H.; Zhou, Y. miR-29c-3p promotes senescence of human mesenchymal stem cells by targeting CNOT6 through p53–p21 and p16–pRB pathways. Biochim. Biophys. Acta 2016, 1863, 520–532. [Google Scholar] [CrossRef]
- Tao, X.; Yang, X.; Wu, K.; Yang, L.; Huang, Y.; Jin, Q.; Chen, S. miR-629–5p promotes growth and metastasis of hepatocellular carcinoma by activating β-catenin. Exp. Cell Res. 2019, 380, 124–130. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Fan, L.; Mou, J.; Yin, Y.; Peng, C.; Chen, Y.; Lu, H.; Zhao, L.; Tao, Z.; et al. MiR-629-5p promotes the invasion of lung adenocarcinoma via increasing both tumor cell invasion and endothelial cell permeability. Oncogene 2020, 39, 3473–3488. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lu, S.; Li, J.; Yu, Q.; Liu, L.; Li, Q. MiR-629-5p promotes colorectal cancer progression through targetting CXXC finger protein 4. Biosci. Rep. 2018, 38, BSR20180613. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Shen, Z.; Qian, H.; Zhou, S.; Chen, Y. Knockdown of miR-629 Inhibits Ovarian Cancer Malignant Behaviors by Targeting Testis-Specific Y-Like Protein 5. DNA Cell Biol. 2017, 36, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, Q.; Wu, J.; Hu, W.; Jiang, J.; Shi, L.; Yang, X.; Zhu, D.; Ji, M.; Wu, C. MiR-629 promotes human pancreatic cancer progression by targeting FOXO3. Cell Death Dis. 2017, 8, e3154. [Google Scholar] [CrossRef]
- Huang, J.; Jiang, R.; Chu, X.; Wang, F.; Sun, X.; Wang, Y.; Pang, L. Overexpression of microRNA -23a-5p induces myocardial infarction by promoting cardiomyocyte apoptosis through inhibited of PI3K / AKT signalling pathway. Cell Biochem. Funct. 2020, 38, 1047–1055. [Google Scholar] [CrossRef]
- Kurkewich, J.L.; Hansen, J.; Klopfenstein, N.; Zhang, H.; Wood, C.; Boucher, A.; Hickman, J.; Muench, D.E.; Grimes, H.L.; Dahl, R. The miR-23a~27a~24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 2017, 13, e1006887. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.-Y.; Feng, Y.-G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in Human Cancer: Its Roles, Mechanisms and Therapeutic Relevance. Cancers 2018, 11, 7. [Google Scholar] [CrossRef]
- Huang, W.; Huang, Y.; Gu, J.; Zhang, J.; Yang, J.; Liu, S.; Xie, C.; Fan, Y.; Wang, H. miR-23a-5p inhibits cell proliferation and invasion in pancreatic ductal adenocarcinoma by suppressing ECM1 expression. Am. J. Transl. Res. 2019, 11, 2983–2994. [Google Scholar]
- Ganesan, S.; Palani, H.K.; Lakshmanan, V.; Balasundaram, N.; Abu Alex, A.; David, S.; Venkatraman, A.; Korula, A.; George, B.; Balasubramanian, P.; et al. Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Quan, J.; Jin, L.; Pan, X.; He, T.; Lai, Y.; Chen, P.; Lin, C.; Yang, S.; Zeng, H.; Lai, Y. Oncogenic miR-23a-5p is associated with cellular function in RCC. Mol. Med. Rep. 2017, 16, 2309–2317. [Google Scholar] [CrossRef]
- Stanczyk, J.; Pedrioli, D.M.; Brentano, F.; Sanchez-Pernaute, O.; Kolling, C.; Gay, R.E.; Detmar, M.; Gay, S.; Kyburz, D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008, 58, 1001–1009. [Google Scholar] [CrossRef]
- Yang, J.-X.; Xie, P.; Li, Y.-S.; Wen, T.; Yang, X.-C. Osteoclast-derived miR-23a-5p-containing exosomes inhibit osteogenic differentiation by regulating Runx2. Cell. Signal. 2019, 70, 109504. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Xue, W.; Liang, J.; Wang, L. MiR-874 Inhibits Cell Proliferation, Migration, and Invasion of Glioma Cells and Correlates with Prognosis of Glioma Patients. NeuroMolecular Med. 2020, 23, 247–255. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Rashed, M.R.; Bayraktar, R.; Mitra, R.; Ivan, C.; Aslan, B.; Zhang, X.; Filant, J.; Silva, A.M.; Rodriguez-Aguayo, C.; et al. Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res. 2016, 76, 7194–7207. [Google Scholar] [CrossRef]



| Total (n = 10) | SGA-nCU | AGA | p-Value |
|---|---|---|---|
| n (%) | 5 (50%) | 5 (50%) | - |
| Boys/girls | 3/2 | 2/3 | - |
| GA (weeks) | 38.80 ± 1.64 | 39.40 ± 1.34 | 0.548 |
| Birth weight (kg) | 2.44 ± 0.18 | 3.34 ± 0.27 | 0.008 |
| CA (year) | 4.54 ± 0.28 | 4.63 ± 0.28 | 0.690 |
| Height (cm) | 97.22 ± 1.96 | 106.94 ± 1.66 | 0.008 |
| Height SDS | –2.17 ± 0.19 | 0.15 ± 0.45 | 0.008 |
| Weight (kg) | 13.62 ± 1.62 | 17.30 ± 1.83 | 0.008 |
| Weight SDS | –2.51 ± 1.01 | –0.28 ± 0.79 | 0.008 |
| BMI SDS | –1.26 ± 1.15 | –0.62 ± 1.15 | 0.690 |
| Total (n = 16) | SGA | AGA | p-Value | |
|---|---|---|---|---|
| SGA-nCU | SGA-CU | |||
| n (%) | 6 (37.5) | 5 (31.25) | 5 (31.25) | - |
| Boys/girls | 3/3 | 2/3 | 2/3 | - |
| GA (weeks) | 38.60 ± 1.36 | 38.20 ± 1.09 | 39.20 ± 1.30 | 0.353 |
| Birth weight (kg) | 2.39 ± 0.24 | 2.06 ± 0.23 | 3.47 ± 0.29 | 0.003 |
| CA (year) | 4.44 ± 0.31 | 5.93 ± 0.77 | 5.87 ± 1.06 | 0.018 |
| Height (cm) | 96.66 ± 1.89 | 111.66 ± 3.78 | 117.58 ± 7.92 | 0.004 |
| Height SDS | –2.10 ± 0.27 | –0.61 ± 0.39 | 0.69 ± 0.82 | 0.001 |
| Weight (kg) | 14.16 ± 1.74 | 17.96 ± 1.88 | 25.50 ± 7.42 | 0.003 |
| Weight SDS | –2.06 ± 1.14 | –1.17 ± 0.67 | 1.19 ± 1.15 | 0.006 |
| BMI SDS | –0.62 ± 1.21 | –1.17 ± 0.85 | 1.12 ± 1.52 | 0.025 |
| miRNA | Fold Change SGA-nCU/AGA | p-Value |
|---|---|---|
| hsa-miR-29b-3p | 7.03 | <0.001 |
| hsa-miR-4448 | 5.34 | 0.000 |
| hsa-miR-141-3p | 4.97 | 0.001 |
| hsa-miR-29a-3p | 4.18 | 0.004 |
| hsa-miR-144-5p | 4.14 | 0.003 |
| hsa-miR-26b-5p | 4.00 | 0.006 |
| hsa-miR-29c-3p | 3.93 | 0.008 |
| hsa-let-7g-5p | 3.81 | 0.002 |
| hsa-miR-32-5p | 3.48 | 0.011 |
| hsa-miR-96-5p | 3.45 | 0.012 |
| hsa-miR-543 | –2.56 | 0.031 |
| hsa-miR-493-3p | –2.64 | 0.002 |
| hsa-miR-125b-1-3p | –2.66 | 0.017 |
| hsa-miR-409-3p | –2.86 | 0.013 |
| hsa-miR-3120-5p | –2.90 | 0.030 |
| hsa-miR-485-3p | –2.92 | 0.032 |
| hsa-miR-758-3p | –2.98 | 0.002 |
| hsa-miR-654-5p | –3.30 | 0.002 |
| hsa-miR-370-3p | –3.35 | 0.005 |
| hsa-miR-127-3p | –3.49 | 0.005 |
| miRNA | Fold Change SGA-nCU/AGA | p-Value |
|---|---|---|
| hsa-miR-3651 | 11.17 | 0.006 |
| hsa-miR-874-3p | 3.65 | 0.002 |
| hsa-miR-363-3p | 3.37 | 0.007 |
| hsa-miR-29a-3p | 2.44 | 0.049 |
| hsa-miR-15a-5p | 2.44 | 0.043 |
| hsa-miR-29c-3p | 2.43 | 0.009 |
| hsa-miR-30c-5p | 2.09 | 0.043 |
| hsa-miR-505-5p | –2.05 | 0.039 |
| hsa-let-7d-5p | –2.07 | 0.044 |
| hsa-miR-629-5p | –2.24 | 0.040 |
| hsa-miR-6126 | –2.35 | 0.049 |
| hsa-let-7f-5p | –2.78 | 0.033 |
| hsa-let-7a-5p | –2.87 | 0.029 |
| hsa-let-7e-5p | –2.93 | 0.007 |
| hsa-miR-23a-5p | –3.25 | 0.004 |
| hsa-miR-28-5p | –4.80 | 0.002 |
| hsa-miR-1-3p | –4.80 | 0.004 |
| (a) | ||||
|---|---|---|---|---|
| miRNA | Fold Change SGA-CU/AGA | p-Value | ||
| hsa-miR-1285-5p | 3.90 | 0.006 | ||
| hsa-miR-874-3p | 3.29 | 0.004 | ||
| hsa-miR-339-3p | 3.06 | 0.027 | ||
| hsa-miR-143-3p | 2.08 | 0.038 | ||
| hsa-miR-6126 | –2.12 | 0.027 | ||
| hsa-miR-627-5p | –4.26 | 0.044 | ||
| (b) | ||||
| SGA-nCU vs. AGA | SGA-CU vs. AGA | |||
| miRNA | Fold Change | p-Value | Fold Change | p-Value |
| hsa-miR-874-3p | 3.65 | 0.002 | 3.29 | 0.004 |
| hsa-miR-6126 | –2.35 | 0.049 | –2.12 | 0.027 |
| (a) | ||||
|---|---|---|---|---|
| miRNA | Fold Change SGA-nCU/SGA-CU | p-Value | ||
| hsa-miR-342-3p | 3.59 | 0.010 | ||
| hsa-miR-576-5p | 3.55 | 0.001 | ||
| hsa-miR-150-5p | 3.10 | 0.006 | ||
| hsa-miR-192-5p | 2.85 | 0.001 | ||
| hsa-miR-29b-3p | 2.83 | 0.006 | ||
| hsa-miR-101-3p | 2.65 | 0.035 | ||
| hsa-miR-30c-5p | 2.58 | 0.010 | ||
| hsa-miR-363-3p | 2.45 | 0.046 | ||
| hsa-miR-29a-3p | 2.38 | 0.037 | ||
| hsa-miR-25-3p | 2.22 | 0.011 | ||
| hsa-miR-21-5p | 2.17 | 0.012 | ||
| hsa-miR-29c-3p | 2.03 | 0.027 | ||
| hsa-miR-335-5p | –2.04 | 0.012 | ||
| hsa-miR-574-5p | –2.05 | 0.035 | ||
| hsa-miR-193a-5p | –2.37 | 0.035 | ||
| hsa-miR-629-5p | –2.62 | 0.012 | ||
| hsa-miR-23a-5p | –2.85 | 0.005 | ||
| hsa-miR-382-5p | –2.88 | 0.039 | ||
| hsa-miR-326 | –2.91 | 0.007 | ||
| (b) | ||||
| SGA-nCU vs. SGA-CU | SGA-nCU vs. AGA | |||
| miRNA | Fold Change | p-Value | Fold Change | p-Value |
| hsa-miR-30c-5p | 2.58 | 0.010 | 2.09 | 0.043 |
| hsa-miR-363-3p | 2.45 | 0.046 | 3.37 | 0.007 |
| hsa-miR-29a-3p | 2.38 | 0.037 | 2.44 | 0.049 |
| hsa-miR-29c-3p | 2.03 | 0.027 | 2.43 | 0.009 |
| hsa-miR-629-5p | –2.62 | 0.012 | –2.24 | 0.040 |
| hsa-miR-23a-5p | –2.85 | 0.005 | –3.25 | 0.004 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.R.; Han, J.-A.; Kim, H.; Lee, H.J.; Shim, Y.S.; Kang, M.J.; Yoon, J.S.; Ryu, S.; Hwang, I.T. Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth. Genes 2022, 13, 938. https://doi.org/10.3390/genes13060938
Jeong HR, Han J-A, Kim H, Lee HJ, Shim YS, Kang MJ, Yoon JS, Ryu S, Hwang IT. Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth. Genes. 2022; 13(6):938. https://doi.org/10.3390/genes13060938
Chicago/Turabian StyleJeong, Hwal Rim, Jae-A Han, Heeji Kim, Hye Jin Lee, Young Suk Shim, Min Jae Kang, Jong Seo Yoon, Seongho Ryu, and Il Tae Hwang. 2022. "Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth" Genes 13, no. 6: 938. https://doi.org/10.3390/genes13060938
APA StyleJeong, H. R., Han, J.-A., Kim, H., Lee, H. J., Shim, Y. S., Kang, M. J., Yoon, J. S., Ryu, S., & Hwang, I. T. (2022). Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth. Genes, 13(6), 938. https://doi.org/10.3390/genes13060938







