miRNAs Copy Number Variations Repertoire as Hallmark Indicator of Cancer Species Predisposition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Statistical Comparison
2.3. Pathways Analysis
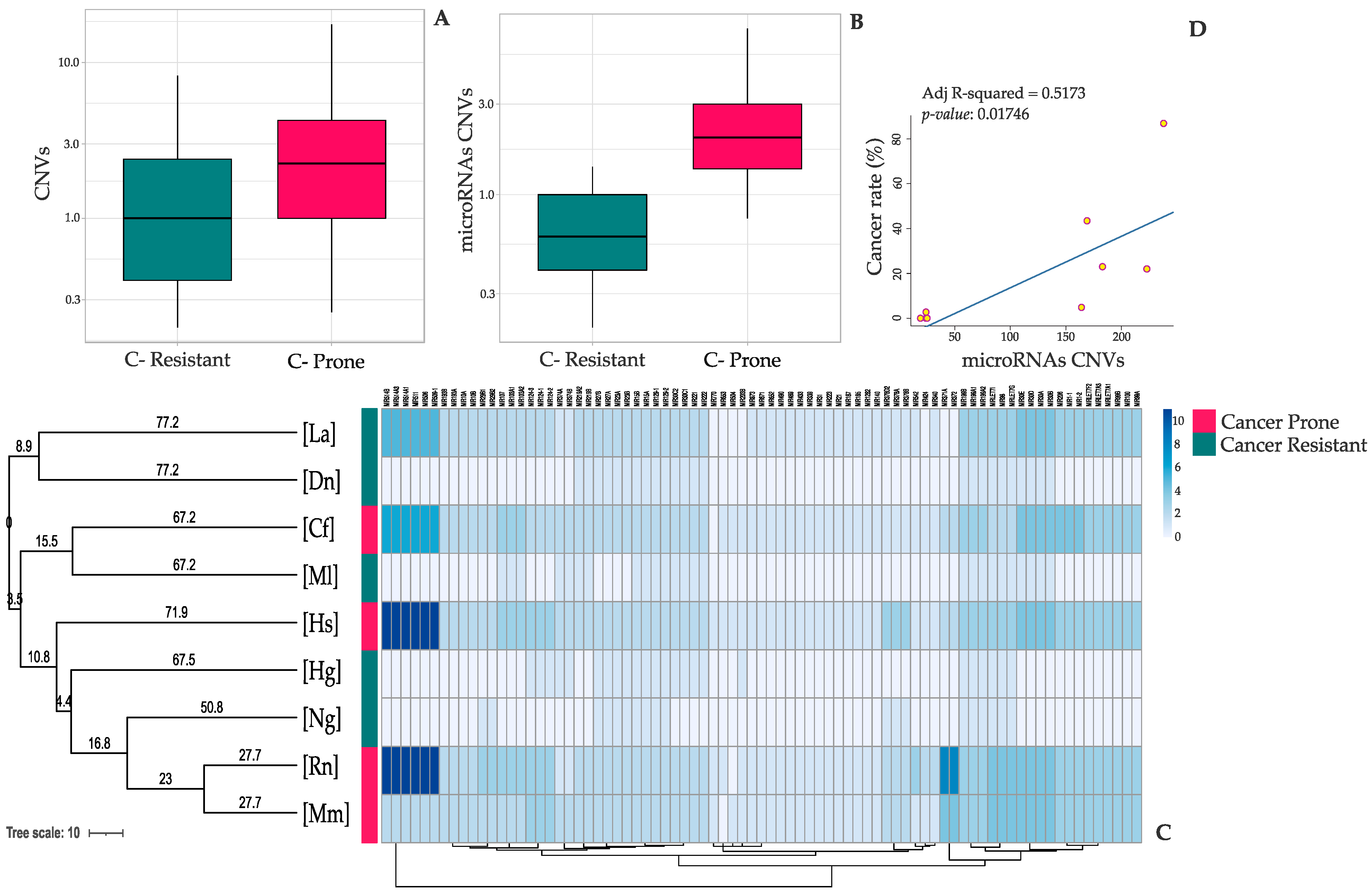
3. Results
3.1. Best Candidate Cancer-Related Genes
3.2. Cancer-Related MicroRNAs Pathways Are among the Most Significantly Enriched Biological Families
3.3. ORA Analysis Confirms a Significant Enrichment in the miRNAs Gene Family
4. Discussion
5. Limitations and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Serrano, M. Unraveling the Links between Cancer and Aging. Carcinogenesis 2016, 37, 107. [Google Scholar] [CrossRef] [PubMed]
- Peto, R.; Roe, F.J.; Lee, P.N.; Levy, L.; Clack, J. Cancer and Ageing in Mice and Men. Br. J. Cancer 1975, 32, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Buffenstein, R. Negligible Senescence in the Longest Living Rodent, the Naked Mole-Rat: Insights from a Successfully Aging Species. J. Comp. Physiol. B 2008, 178, 439–445. [Google Scholar] [CrossRef]
- Liang, S.; Mele, J.; Wu, Y.; Buffenstein, R.; Hornsby, P.J. Resistance to Experimental Tumorigenesis in Cells of a Long-Lived Mammal, the Naked Mole-Rat (Heterocephalus Glaber): Oncogene Resistance in Naked Mole-Rat Cells. Aging Cell 2010, 9, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.B.; Fang, X.; Fushan, A.A.; Huang, Z.; Lobanov, A.V.; Han, L.; Marino, S.M.; Sun, X.; Turanov, A.A.; Yang, P.; et al. Genome Sequencing Reveals Insights into Physiology and Longevity of the Naked Mole Rat. Nature 2011, 479, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Seluanov, A.; Hine, C.; Azpurua, J.; Feigenson, M.; Bozzella, M.; Mao, Z.; Catania, K.C.; Gorbunova, V. Hypersensitivity to Contact Inhibition Provides a Clue to Cancer Resistance of Naked Mole-Rat. Proc. Natl. Acad. Sci. USA 2009, 106, 19352–19357. [Google Scholar] [CrossRef]
- Lipman, R.; Galecki, A.; Burke, D.T.; Miller, R.A. Genetic Loci That Influence Cause of Death in a Heterogeneous Mouse Stock. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, B977–B983. [Google Scholar] [CrossRef]
- Boddy, A.M.; Harrison, T.M.; Abegglen, L.M. Comparative Oncology: New Insights into an Ancient Disease. iScience 2020, 23, 101373. [Google Scholar] [CrossRef]
- Wilkinson, G.S.; Adams, D.M. Recurrent Evolution of Extreme Longevity in Bats. Biol. Lett. 2019, 15, 20180860. [Google Scholar] [CrossRef]
- Tollis, M.; Schiffman, J.D.; Boddy, A.M. Evolution of Cancer Suppression as Revealed by Mammalian Comparative Genomics. Curr. Opin. Genet. Dev. 2017, 42, 40–47. [Google Scholar] [CrossRef]
- Wang, L.-F.; Walker, P.J.; Poon, L.L.M. Mass Extinctions, Biodiversity and Mitochondrial Function: Are Bats ‘Special’ as Reservoirs for Emerging Viruses? Curr. Opin. Virol. 2011, 1, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Abegglen, L.M.; Caulin, A.F.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.S.; Kiso, W.K.; Schmitt, D.L.; Waddell, P.J.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012: Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Tian, X.; Seluanov, A.; Gorbunova, V. Molecular Mechanisms Determining Lifespan in Short- and Long-Lived Species. Trends Endocrinol. Metab. 2017, 28, 722–734. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.P.; Emes, R.D.; Lynch, V.J. TP53 Copy Number Expansion Is Associated with the Evolution of Increased Body Size and an Enhanced DNA Damage Response in Elephants. eLife 2016, 5, e11994. [Google Scholar] [CrossRef] [PubMed]
- Caulin, A.F.; Maley, C.C. Peto’s Paradox: Evolution’s Prescription for Cancer Prevention. Trends Ecol. Evol. 2011, 26, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural Variation in the Human Genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The Cancer Genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Tollis, M.; Boddy, A.M.; Maley, C.C. Peto’s Paradox: How Has Evolution Solved the Problem of Cancer Prevention? BMC Biol. 2017, 15, 60. [Google Scholar] [CrossRef]
- Tollis, M.; Schneider-Utaka, A.K.; Maley, C.C. The Evolution of Human Cancer Gene Duplications across Mammals. Mol. Biol. Evol. 2020, 37, 2875–2886. [Google Scholar] [CrossRef]
- Boddy, A.M.; Abegglen, L.M.; Pessier, A.P.; Aktipis, A.; Schiffman, J.D.; Maley, C.C.; Witte, C. Lifetime Cancer Prevalence and Life History Traits in Mammals. Evol. Med. Public Health 2020, 2020, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The Landscape of Somatic Copy-Number Alteration across Human Cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Bignell, G.R.; Greenman, C.D.; Davies, H.; Butler, A.P.; Edkins, S.; Andrews, J.M.; Buck, G.; Chen, L.; Beare, D.; Latimer, C.; et al. Signatures of Mutation and Selection in the Cancer Genome. Nature 2010, 463, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Iskow, R.C.; Gokcumen, O.; Lee, C. Exploring the Role of Copy Number Variants in Human Adaptation. Trends Genet. 2012, 28, 245–257. [Google Scholar] [CrossRef]
- Vischioni, C.; Bove, F.; Mandreoli, F.; Martoglia, R.; Pisi, V.; Taccioli, C. Visual Exploratory Data Analysis for Copy Number Variation Studies in Biomedical Research. Big Data Res. 2022, 27, 100298. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Herrero, J.; Muffato, M.; Beal, K.; Fitzgerald, S.; Gordon, L.; Pignatelli, M.; Vilella, A.J.; Searle, S.M.J.; Amode, R.; Brent, S.; et al. Ensembl Comparative Genomics Resources. Database 2016, 2016, 17. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A Computational Tool for the Study of Gene Family Evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Seluanov, A.; Gladyshev, V.N.; Vijg, J.; Gorbunova, V. Mechanisms of Cancer Resistance in Long-Lived Mammals. Nat. Rev. Cancer 2018, 18, 433–441. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. Cancer-Free Aging: Insights from Spalax Ehrenbergi Superspecies. Ageing Res. Rev. 2018, 47, 18–23. [Google Scholar] [CrossRef]
- Ma, S.; Gladyshev, V.N. Molecular Signatures of Longevity: Insights from Cross-Species Comparative Studies. Semin. Cell Dev. Biol. 2017, 70, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Hine, C.; Tian, X.; Ablaeva, J.; Gudkov, A.V.; Nevo, E.; Seluanov, A. Cancer Resistance in the Blind Mole Rat Is Mediated by Concerted Necrotic Cell Death Mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 19392–19396. [Google Scholar] [CrossRef] [PubMed]
- Shepard, A.; Kissil, J.L. The Use of Non-Traditional Models in the Study of Cancer Resistance—The Case of the Naked Mole Rat. Oncogene 2020, 39, 5083–5097. [Google Scholar] [CrossRef] [PubMed]
- Orme, D. The Caper Package: Comparative Analysis of Phylogenetics and Evolution in R. 36. Available online: https://cran.r-project.org/web/packages/caper/vignettes/caper.pdf (accessed on 4 May 2022).
- Upham, N.S.; Esselstyn, J.A.; Jetz, W. Inferring the Mammal Tree: Species-Level Sets of Phylogenies for Questions in Ecology, Evolution, and Conservation. PLOS Biol. 2019, 17, e3000494. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Postma, M.; Goedhart, J. PlotsOfData—A Web App for Visualizing Data Together with Their Summaries. PLOS Biol. 2019, 17, e3000202. [Google Scholar] [CrossRef]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A Web Tool for Generation of Box Plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E.; et al. MiRTargetLink 2.0—Interactive MiRNA Target Gene and Target Pathway Networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. PLoS Comput. Biol. 2012, 8, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, G.; Sui, Y.; Yang, Z.; Chu, Y.; Tang, H.; Guo, B.; Zhang, C.; Wu, C. CD52 Is a Prognostic Biomarker and Associated with Tumor Microenvironment in Breast Cancer. Front. Genet. 2020, 11, 578002. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.S.; Aguila, B.; Brett-Morris, A.; Creighton, C.J.; Welford, S.M. Spermidine/Spermine N1-Acetyltransferase 1 Is a Gene-Specific Transcriptional Regulator That Drives Brain Tumor Aggressiveness. Oncogene 2019, 38, 6794–6800. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, F.; Wang, X.; Cheng, B.; Ye, F.; Xie, X.; Zhou, C.; Lu, W. Suppressed MiR-424 Expression via Upregulation of Target Gene Chk1 Contributes to the Progression of Cervical Cancer. Oncogene 2013, 32, 976–987. [Google Scholar] [CrossRef]
- Fan, X.; Huang, X.; Li, Z.; Ma, X. MicroRNA-372-3p Promotes the Epithelial-Mesenchymal Transition in Breast Carcinoma by Activating the Wnt Pathway. J. BUON 2018, 23, 1309–1315. [Google Scholar]
- Sun, H.; Gao, D. Propofol Suppresses Growth, Migration and Invasion of A549 Cells by down-Regulation of MiR-372. BMC Cancer 2018, 18, 1252. [Google Scholar] [CrossRef]
- Jones, L.; Naidoo, M.; Machado, L.R.; Anthony, K. The Duchenne Muscular Dystrophy Gene and Cancer. Cell. Oncol. 2021, 44, 19–32. [Google Scholar] [CrossRef]
- Spilka, R.; Ernst, C.; Mehta, A.K.; Haybaeck, J. Eukaryotic Translation Initiation Factors in Cancer Development and Progression. Cancer Lett. 2013, 340, 9–21. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Lotterman, C.D.; Bao, C.; Hruban, R.H.; Karim, B.; Mendell, J.T.; Huso, D.; Lowenstein, C.J. P53-Induced MicroRNA-107 Inhibits HIF-1 and Tumor Angiogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6334–6339. [Google Scholar] [CrossRef]
- Turco, C.; Donzelli, S.; Fontemaggi, G. MiR-15/107 MicroRNA Gene Group: Characteristics and Functional Implications in Cancer. Front. Cell Dev. Biol. 2020, 8, 427. [Google Scholar] [CrossRef]
- Wang, P.; Chen, L.; Zhang, J.; Chen, H.; Fan, J.; Wang, K.; Luo, J.; Chen, Z.; Meng, Z.; Liu, L. Methylation-Mediated Silencing of the MiR-124 Genes Facilitates Pancreatic Cancer Progression and Metastasis by Targeting Rac1. Oncogene 2014, 33, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Ledl, A.; Schmidt, D. SUMO: A Regulator of Gene Expression and Genome Integrity. Oncogene 2004, 23, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Schneeweis, C.; Hassan, Z.; Schick, M.; Keller, U.; Schneider, G. The SUMO Pathway in Pancreatic Cancer: Insights and Inhibition. Br. J. Cancer 2021, 124, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.-Y.; Lin, Y.; Yu, Y.-Q.; Cao, S.-J.; Zhang, R.; Yang, X.-M.; Li, J.; Zhang, Y.-L.; Wang, Y.-H.; Ma, M.-Z.; et al. MiR-506 Acts as a Tumor Suppressor by Directly Targeting the Hedgehog Pathway Transcription Factor Gli3 in Human Cervical Cancer. Oncogene 2015, 34, 717–725. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhou, L.; Zhao, C.; Wan, J.; Yu, Z.; Guo, X.; Qin, J.; Chen, J.; Lu, R. Identification of MiR-508-3p and MiR-509-3p That Are Associated with Cell Invasion and Migration and Involved in the Apoptosis of Renal Cell Carcinoma. Biochem. Biophys. Res. Commun. 2012, 419, 621–626. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Pucci, F.; Magri, L.; Moi, D.; Gilfillan, G.D.; Ranghetti, A.; Casazza, A.; Mazzone, M.; Lyle, R.; Naldini, L.; et al. MiR-511-3p Modulates Genetic Programs of Tumor-Associated Macrophages. Cell Rep. 2012, 1, 141–154. [Google Scholar] [CrossRef]
- Ren, L.-L.; Yan, T.-T.; Shen, C.-Q.; Tang, J.-Y.; Kong, X.; Wang, Y.-C.; Chen, J.; Liu, Q.; He, J.; Zhong, M.; et al. The Distinct Role of Strand-Specific MiR-514b-3p and MiR-514b-5p in Colorectal Cancer Metastasis. Cell Death Dis. 2018, 9, 687. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, W.; Han, T.; Du, S.; Li, Z.; Zhang, Z.; Shan, G.; Kong, C. MiR-378 Suppresses Prostate Cancer Cell Growth through Downregulation of MAPK1 in Vitro and in Vivo. Tumor Biol. 2016, 37, 2095–2103. [Google Scholar] [CrossRef]
- Zhu, W.; Xue, Y.; Liang, C.; Zhang, R.; Zhang, Z.; Li, H.; Su, D.; Liang, X.; Zhang, Y.; Huang, Q.; et al. S100A16 Promotes Cell Proliferation and Metastasis via AKT and ERK Cell Signaling Pathways in Human Prostate Cancer. Tumor Biol. 2016, 37, 12241–12250. [Google Scholar] [CrossRef]
- Miremadi, A.; Oestergaard, M.Z.; Pharoah, P.D.P.; Caldas, C. Cancer Genetics of Epigenetic Genes. Hum. Mol. Genet. 2007, 16, R28–R49. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, M.; Hu, Q.; Xu, W.; Liu, W.; Ye, Z.; Fan, G.; Xu, X.; Yu, X.; Ji, S.; et al. FGFBP1, a Downstream Target of the FBW7/c-Myc Axis, Promotes Cell Proliferation and Migration in Pancreatic Cancer. Am. J. Cancer Res. 2019, 9, 2650. [Google Scholar] [PubMed]
- Xian, S.; Shang, D.; Kong, G.; Tian, Y. FOXJ1 Promotes Bladder Cancer Cell Growth and Regulates Warburg Effect. Biochem. Biophys. Res. Commun. 2018, 495, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Nohata, N.; Sone, Y.; Hanazawa, T.; Fuse, M.; Kikkawa, N.; Yoshino, H.; Chiyomaru, T.; Kawakami, K.; Enokida, H.; Nakagawa, M.; et al. MiR-1 as a Tumor Suppressive MicroRNA Targeting TAGLN2 in Head and Neck Squamous Cell Carcinoma. Oncotarget 2011, 2, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Jin, H.; Guo, X.; Xia, L.; Chen, Z.; Bai, M.; Liu, J.; Shang, X.; Wu, K.; et al. MiR-206 Inhibits Gastric Cancer Proliferation in Part by Repressing CyclinD2. Cancer Lett. 2013, 332, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wu, Q.; Wang, C.; Wang, X.; Huang, J.; Zhao, J.; Mao, S.; Zhang, G.; Xu, X.; Zhang, N. MiR-340 Inhibition of Breast Cancer Cell Migration and Invasion through Targeting of Oncoprotein c-Met. Cancer 2011, 117, 2842–2852. [Google Scholar] [CrossRef] [PubMed]
- Kureel, J.; Dixit, M.; Tyagi, A.M.; Mansoori, M.N.; Srivastava, K.; Raghuvanshi, A.; Maurya, R.; Trivedi, R.; Goel, A.; Singh, D. MiR-542-3p Suppresses Osteoblast Cell Proliferation and Differentiation, Targets BMP-7 Signaling and Inhibits Bone Formation. Cell Death Dis. 2014, 5, e1050. [Google Scholar] [CrossRef] [PubMed]
- Cano, C.E.; Hamidi, T.; Sandi, M.J.; Iovanna, J.L. Nupr1: The Swiss-Knife of Cancer. J. Cell. Physiol. 2011, 226, 1439–1443. [Google Scholar] [CrossRef]
- Peters, K.M.; Carlson, B.A.; Gladyshev, V.N.; Tsuji, P.A. Selenoproteins in Colon Cancer. Free Radic. Biol. Med. 2018, 127, 14–25. [Google Scholar] [CrossRef]
- Yim, S.H.; Clish, C.B.; Gladyshev, V.N. Selenium Deficiency Is Associated with Pro-Longevity Mechanisms. Cell Rep. 2019, 27, 2785–2797.e3. [Google Scholar] [CrossRef]
- Elliott, B.; Millena, A.C.; Matyunina, L.; Zhang, M.; Zou, J.; Wang, G.; Zhang, Q.; Bowen, N.; Eaton, V.; Webb, G.; et al. Essential Role of JunD in Cell Proliferation Is Mediated via MYC Signaling in Prostate Cancer Cells. Cancer Lett. 2019, 448, 155–167. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Miller, R.A.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting Communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kourtidis, A.; Lu, R.; Pence, L.J.; Anastasiadis, P.Z. A Central Role for Cadherin Signaling in Cancer. Exp. Cell Res. 2017, 358, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, J.M.; Lynch, V.J. Pervasive Duplication of Tumor Suppressors in Afrotherians during the Evolution of Large Bodies and Reduced Cancer Risk. eLife 2021, 10, e65041. [Google Scholar] [CrossRef] [PubMed]
- Glenfield, C.; Innan, H. Gene Duplication and Gene Fusion Are Important Drivers of Tumourigenesis during Cancer Evolution. Genes 2021, 12, 1376. [Google Scholar] [CrossRef]
- Eifler, K.; Vertegaal, A.C.O. SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem. Sci. 2015, 40, 779–793. [Google Scholar] [CrossRef]
- Gallia, G.L.; Zhang, M.; Ning, Y.; Haffner, M.C.; Batista, D.; Binder, Z.A.; Bishop, J.A.; Hann, C.L.; Hruban, R.H.; Ishii, M.; et al. Genomic Analysis Identifies Frequent Deletions of Dystrophin in Olfactory Neuroblastoma. Nat. Commun. 2018, 9, 5410. [Google Scholar] [CrossRef]
- Ruggieri, S.; De Giorgis, M.; Annese, T.; Tamma, R.; Notarangelo, A.; Marzullo, A.; Senetta, R.; Cassoni, P.; Notarangelo, M.; Ribatti, D.; et al. Dp71 Expression in Human Glioblastoma. Int. J. Mol. Sci. 2019, 20, 5429. [Google Scholar] [CrossRef]
- Schmiedel, J.M.; Klemm, S.L.; Zheng, Y.; Sahay, A.; Blüthgen, N.; Marks, D.S.; van Oudenaarden, A. MicroRNA Control of Protein Expression Noise. Science 2015, 348, 128–132. [Google Scholar] [CrossRef]
- Martinez-Sanchez, A.; Murphy, C. MicroRNA Target Identification—Experimental Approaches. Biology 2013, 2, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.D.; Lund, A.H. MicroRNA and Cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. MiR-30b/30d Regulation of GalNAc Transferases Enhances Invasion and Immunosuppression during Metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. MiR221/222 in Cancer: Their Role in Tumor Progression and Response to Therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.V.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA Cluster 221-222 and Estrogen Receptor α Interactions in Breast Cancer. JNCI J. Natl. Cancer Inst. 2010, 102, 706–721. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and MiR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. BCL2 and MiR-15/16: From Gene Discovery to Treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef]
- Tang, Y.-C.; Amon, A. Gene Copy-Number Alterations: A Cost-Benefit Analysis. Cell 2013, 152, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.J.; Resio, B.; Pellman, D. Causes and Consequences of Aneuploidy in Cancer. Nat. Rev. Genet. 2012, 13, 189–203. [Google Scholar] [CrossRef]
- Kirkwood, T.B.L. Why and How Are We Living Longer?: Why and How Are We Living Longer? Exp. Physiol. 2017, 102, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Holtze, S.; Gorshkova, E.; Braude, S.; Cellerino, A.; Dammann, P.; Hildebrandt, T.B.; Hoeflich, A.; Hoffmann, S.; Koch, P.; Terzibasi Tozzini, E.; et al. Alternative Animal Models of Aging Research. Front. Mol. Biosci. 2021, 8, 660959. [Google Scholar] [CrossRef] [PubMed]
- Hajibabaie, F.; Abedpoor, N.; Assareh, N.; Tabatabaiefar, M.A.; Shariati, L.; Zarrabi, A. The Importance of SNPs at MiRNA Binding Sites as Biomarkers of Gastric and Colorectal Cancers: A Systematic Review. J. Pers. Med. 2022, 12, 456. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Non-Coding RNA 2021, 7, 47. [Google Scholar] [CrossRef] [PubMed]

| Gene | p-Value | Known_TS | Known_OG | References |
|---|---|---|---|---|
| CD52 | 0.007 | NO | NO | [43] |
| SAT1 | 0.007 | NO | NO | [44] |
| MIR424 | 0.009 | YES | NO | [45] |
| MIR372 | 0.010 | NO | YES | [46,47] |
| DMD | 0.014 | YES | NO | [48] |
| EIF5 | 0.017 | NO | NO | [49] |
| MIR107 | 0.022 | YES | YES | [50,51] |
| MIR124-1, MIR124-2, MIR124-3 | 0.022 | YES | NO | [52] |
| SUMO2, SUMO3, SUMO4 | 0.024 | NO | NO | [53,54] |
| MIR506 | 0.029 | YES | YES | [55] |
| MIR509-1 | 0.029 | NO | NO | [56] |
| MIR511 | 0.029 | YES | NO | [57] |
| MIR514A1, MIR514A3, MIR514B | 0.029 | NO | NO | [58] |
| MIR378A | 0.030 | YES | NO | [59] |
| S100A16 | 0.030 | NO | NO | [60] |
| MBD1, MBD2, MBD3 | 0.031 | NO | YES (MDB1) | [61] |
| FGFBP1 | 0.032 | NO | NO | [62] |
| FOXJ1 | 0.032 | NO | NO | [63] |
| MIR1-1, MIR1-2 | 0.032 | YES | NO | [64] |
| MIR206 | 0.032 | YES | NO | [65] |
| MIR340 | 0.032 | YES | NO | [66] |
| MIR542 | 0.032 | NO | NO | [67] |
| NUPR1 | 0.032 | YES | NO | [68] |
| SELENOW | 0.032 | NO | NO | [69,70] |
| JUND | 0.034 | NO | YES | [71] |
| Description | FDR (BH) | Genes | |
|---|---|---|---|
| KEGG | MicroRNAs in cancer | 0 | MIR103A1; MIR103A2; MIR107; MIR124-1; MIR124-2; MIR124-3; MIR1-1; MIR1-2; MIR206; MIR100; MIR10A; MIR10B; MIR129-1; MIR129-2; MIR15A; MIR15B; MIR193B; MIR199A1; MIR199A2; MIR199B; MIR203B; MIR21; MIR223; MIR31; MIR99A; MIRLET7A1; MIRLET7A3; MIRLET7F2; MIR29B1; MIR29B2; MIRLET7G; MIRLET7I; MIR221; MIR222; MIR23A; MIR23B; MIR27A; MIR27B; MIR30C1; MIR30C2; MIR30A; MIR30B; MIR30D; MIR30E. |
| Taste transduction | 3.16 × 10−10 | TAS2R10; TAS2R13; TAS2R14; TAS2R19; TAS2R20; TAS2R3; TAS2R30; TAS2R31; TAS2R42; TAS2R43; TAS2R45; TAS2R46; TAS2R50; TAS2R7; TAS2R8; TAS2R9 | |
| Progesterone-mediated oocyte maturation | 2.43 × 10−4 | SPDYE1; SPDYE11; SPDYE16; SPDYE17; SPDYE2; SPDYE2B; SPDYE3; SPDYE4; SPDYE5; SPDYE6; INS | |
| Oocyte meiosis | 2.73 × 10−4 | PPP3R2; SPDYE1; SPDYE11; SPDYE16; SPDYE17; SPDYE2; SPDYE2B; SPDYE3; SPDYE4; SPDYE5; SPDYE6; INS | |
| PANTHER | Cadherin signaling pathway | 4.02 × 10−2 | PCDHB14; PCDHB7; PCDHGB1; PCDHB16; PCDHB6; PCDHGB4; PCDHGA6; PCDHGB6; PCDHGB7 |
| Wikipathway | miRNAs involved in DNA damage response | 3.76 × 10−9 | MIR371A; MIR372; MIR542; MIR100; MIR15B; MIRLET7A1; MIR374B; MIR221; MIR222; MIR23A; MIR23B; MIR27A; MIR27B |
| Alzheimers Disease | 5.31 × 10−5 | MIR124-1; MIR124-2; MIR124-3; MIR10A; MIR129-1; MIR129-2; MIR199B; MIR21; MIR433; MIR671; MIR873; PPP3R2; MIR29B1; MIR30C2; MIR219A2 | |
| Metastatic brain tumor | 2.31 × 10−3 | MIRLET7A1; MIRLET7A3; MIRLET7F2; MIR29B1; MIR29B2; MIRLET7G | |
| miRNA targets in ECM and membrane receptors | 2.31 × 10−3 | MIR107; MIR15B; MIR30C1; MIR30C2; MIR30B; MIR30D; MIR30E | |
| MicroRNAs in cardiomyocyte hypertrophy | 2.77 × 10−3 | MIR103A1; MIR103A2; MIR140; MIR15B; MIR185; MIR199A1; MIR199A2; MIR23A; MIR27B; MIR30E | |
| Cell Differentiation - Index | 1.25 × 10−2 | MIR1-1; MIR206; MIR199A1; MIR199A2; MIR221; MIR222 | |
| let-7 inhibition of ES cell reprogramming | 1.25 × 10−2 | MIRLET7A1; MIRLET7F2; MIRLET7G; MIRLET7I | |
| miRNAs involvement in the immune response in sepsis | 1.43 × 10−2 | MIR187; MIR199A1; MIR199A2; MIR203B; MIR223; MIR29B1; MIRLET7I | |
| Cell Differentiation-Index expanded | 2.38 × 10−2 | MIR1-1; MIR206; MIR199A1; MIR199A2; MIR221; MIR222 | |
| Role of Osx and miRNAs in tooth development | 3.35 × 10−2 | MIRLET7A1; MIRLET7F2; MIR29B1; MIRLET7G; MIRLET7I |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vischioni, C.; Bove, F.; De Chiara, M.; Mandreoli, F.; Martoglia, R.; Pisi, V.; Liti, G.; Taccioli, C. miRNAs Copy Number Variations Repertoire as Hallmark Indicator of Cancer Species Predisposition. Genes 2022, 13, 1046. https://doi.org/10.3390/genes13061046
Vischioni C, Bove F, De Chiara M, Mandreoli F, Martoglia R, Pisi V, Liti G, Taccioli C. miRNAs Copy Number Variations Repertoire as Hallmark Indicator of Cancer Species Predisposition. Genes. 2022; 13(6):1046. https://doi.org/10.3390/genes13061046
Chicago/Turabian StyleVischioni, Chiara, Fabio Bove, Matteo De Chiara, Federica Mandreoli, Riccardo Martoglia, Valentino Pisi, Gianni Liti, and Cristian Taccioli. 2022. "miRNAs Copy Number Variations Repertoire as Hallmark Indicator of Cancer Species Predisposition" Genes 13, no. 6: 1046. https://doi.org/10.3390/genes13061046
APA StyleVischioni, C., Bove, F., De Chiara, M., Mandreoli, F., Martoglia, R., Pisi, V., Liti, G., & Taccioli, C. (2022). miRNAs Copy Number Variations Repertoire as Hallmark Indicator of Cancer Species Predisposition. Genes, 13(6), 1046. https://doi.org/10.3390/genes13061046








