NUMTs Can Imitate Biparental Transmission of mtDNA—A Case in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crosses
2.2. DNA Extraction and PCR Assays
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Bensasson, D.; Zhang, D.X.; Hartl, D.L.; Hewitt, G.M. Mitochondrial Pseudogenes: Evolution’s Misplaced Witnesses. Trends Ecol. Evol. 2001, 16, 314–321. [Google Scholar] [CrossRef]
- Ricchetti, M.; Tekaia, F.; Dujon, B. Continued Colonization of the Human Genome by Mitochondrial DNA. PLoS Biol. 2004, 2, e273. [Google Scholar] [CrossRef]
- Kowal, K.; Tkaczyk, A.; Pierzchała, M.; Bownik, A.; Ślaska, B. Identification of Mitochondrial DNA (NUMTs) in the Nuclear Genome of Daphnia Magna. Int. J. Mol. Sci. 2020, 21, 8725. [Google Scholar] [CrossRef]
- Laine, V.N.; Gossmann, T.I.; Van Oers, K.; Visser, M.E.; Groenen, M.A.M. Exploring the Unmapped DNA and RNA Reads in a Songbird Genome. BMC Genom. 2019, 20, 19. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-X.; Liu, J.; Miao, Y.-H.; Huang, D.-W.; Xiao, J.-H. Tracking the Distribution and Burst of Nuclear Mitochondrial DNA Sequences (NUMTs) in Fig Wasp Genomes. Insects 2020, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Jiang, F.; Yuan, J.; Guo, W.; Miao, Y. Exceptionally High Cumulative Percentage of NUMTs Originating from Linear Mitochondrial DNA Molecules in the Hydra Magnipapillata Genome. BMC Genom. 2013, 14, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, H.H.; Griffiths-Jones, S. Mitochondrial Pseudogenes in the Nuclear Genomes of Drosophila. PLoS ONE 2012, 7, e32593. [Google Scholar] [CrossRef] [Green Version]
- Baldo, L.; de Queiroz, A.; Hedin, M.; Hayashi, C.Y.; Gatesy, J. Nuclear–Mitochondrial Sequences as Witnesses of Past Interbreeding and Population Diversity in the Jumping Bristletail Mesomachilis. Mol. Biol. Evol. 2011, 28, 195–210. [Google Scholar] [CrossRef]
- Hazkani-Covo, E.; Zeller, R.M.; Martin, W. Molecular Poltergeists: Mitochondrial DNA Copies (Numts) in Sequenced Nuclear Genomes. PLoS Genet. 2010, 6, e1000834. [Google Scholar] [CrossRef]
- Kaya, S.; Çıplak, B. Possibility of Numt Co-Amplification from Gigantic Genome of Orthoptera: Testing Efficiency of Standard PCR Protocol in Producing Orthologous COI Sequences. Heliyon 2018, 4, e00929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, R.J.; Ruiz-Ruano, F.J.; Thomas, C.J.E.; Pérez-Ruiz, M.; Jiménez-Bartolomé, M.; Liu, S.; de la Torre, J.; Bella, J.L. Mind the Numt: Finding Informative Mitochondrial Markers in a Giant Grasshopper Genome. J. Zool. Syst. Evol. Res. 2021, 59, 635–645. [Google Scholar] [CrossRef]
- Pamilo, P.; Viljakainen, L.; Vihavainen, A. Exceptionally High Density of NUMTs in the Honeybee Genome. Mol. Biol. Evol. 2007, 24, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Richly, E.; Leister, D. NUMTs in Sequenced Eukaryotic Genomes. Mol. Biol. Evol. 2004, 21, 1081–1084. [Google Scholar] [CrossRef] [Green Version]
- Calvignac, S.; Konecny, L.; Malard, F.; Douaby, C. Preventing the Pollution of Mitochondrial Datasets with Nuclear Mitochondrial Paralogs (Numts). Mitochondrion 2011, 11, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Xing, Y.; Mao, X. The Little Brown Bat Nuclear Genome Contains an Entire Mitochondrial Genome: Real or Artifact? Gene 2017, 629, 64–67. [Google Scholar] [CrossRef]
- Nacer, D.F.; Raposo do Amaral, F. Striking Pseudogenization in Avian Phylogenetics: Numts Are Large and Common in Falcons. Mol. Phylogenet. Evol. 2017, 115, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Verscheure, S.; Backeljau, T.; Desmyter, S. In Silico Discovery of a Nearly Complete Mitochondrial Genome Numt in the Dog (Canis lupus familiaris) Nuclear Genome. Genetica 2015, 143, 453–458. [Google Scholar] [CrossRef]
- Wang, B.; Zhou, X.; Shi, F.; Liu, Z.; Roos, C.; Garber, P.A.; Li, M.; Pan, H. Full-Length Numt Analysis Provides Evidence for Hybridization between the Asian Colobine Genera Trachypithecus and Semnopithecus. Am. J. Primatol. 2015, 77, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Lutz-Bonengel, S.; Niederstätter, H.; Naue, J.; Koziel, R.; Yang, F.; Sänger, T.; Huber, G.; Berger, C.; Pflugradt, R.; Strobl, C.; et al. Evidence for Multi-Copy Mega-NUMT s in the Human Genome. Nucleic Acids Res. 2021, 49, 1517–1531. [Google Scholar] [CrossRef]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental Inheritance of Mitochondrial DNA in Humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Reply to Lutz-Bonengel et Al.: Biparental MtDNA Transmission Is Unlikely to Be the Result of Nuclear Mitochondrial DNA Segments. Proc. Natl. Acad. Sci. USA 2019, 116, 1823–1824. [Google Scholar] [CrossRef] [Green Version]
- Balciuniene, J.; Balciunas, D. A Nuclear MtDNA Concatemer (Mega-NUMT) Could Mimic Paternal Inheritance of Mitochondrial Genome. Front. Genet. 2019, 10, 2018–2020. [Google Scholar] [CrossRef] [PubMed]
- Lutz-Bonengel, S.; Parson, W. No Further Evidence for Paternal Leakage of Mitochondrial DNA in Humans Yet. Proc. Natl. Acad. Sci. USA 2019, 116, 1821–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mossman, J.A.; Biancani, L.M.; Zhu, C.-T.; Rand, D.M. Mitonuclear Epistasis for Development Time and Its Modification by Diet in Drosophila. Genetics 2016, 203, 463–484. [Google Scholar] [CrossRef] [Green Version]
- Montooth, K.L.; Meiklejohn, C.D.; Abt, D.N.; Rand, D.M. Mitochondrial-Nuclear Epistasis Affects Fitness within Species but Does Not Contribute to Fixed Incompatibilities between Species of Drosophila. Evolution 2010, 64, 3364–3379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solignac, M.; Monnerot, M.; Mounolou, J.C. Mitochondrial DNA Evolution in the Melanogaster Species Subgroup of Drosophila. J. Mol. Evol. 1986, 23, 31–40. [Google Scholar] [CrossRef]
- Ashburner, M.; Golic, K.G.; Hawley, R.S. Drosophila: A Laboratory Handbook, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2005; ISBN 9780879697068. [Google Scholar]
- O’neill, S.L.; Giordano, R.; Colbert, A.M.E.; Karrf, T.L.; Robertson, H.M. 16S RRNA Phylogenetic Analysis of the Bacterial Endosymbionts Associated with Cytoplasmic Incompatibility in Insects. Proc. Natl. Acad. Sci. USA 1992, 89, 2699–2702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [Green Version]
- Dokianakis, E.; Ladoukakis, E.D. Different Degree of Paternal MtDNA Leakage between Male and Female Progeny in Interspecific Drosophila Crosses. Ecol. Evol. 2014, 4, 2633–2641. [Google Scholar] [CrossRef] [PubMed]
- Polovina, E.S.; Parakatselaki, M.E.; Ladoukakis, E.D. Paternal Leakage of Mitochondrial DNA and Maternal Inheritance of Heteroplasmy in Drosophila Hybrids. Sci. Rep. 2020, 10, 2599. [Google Scholar] [CrossRef]
- Zhu, C.T.; Ingelmo, P.; Rand, D.M. GxGxE for Lifespan in Drosophila: Mitochondrial, Nuclear, and Dietary Interactions That Modify Longevity. PLoS Genet. 2014, 10, e1004354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bevers, R.P.J.; Litovchenko, M.; Kapopoulou, A.; Braman, V.S.; Robinson, M.R.; Auwerx, J.; Hollis, B.; Deplancke, B. Mitochondrial Haplotypes Affect Metabolic Phenotypes in the Drosophila Genetic Reference Panel. Nat. Metab. 2019, 1, 1226–1242. [Google Scholar] [CrossRef] [PubMed]
- Wolfner, M.F. The Gifts That Keep on Giving: Physiological Functions and Evolutionary Dynamics of Male Seminal Proteins in Drosophila. Heredity 2002, 88, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; O’Farrell, P.H. Selections That Isolate Recombinant Mitochondrial Genomes in Animals. eLife 2015, 4, e07247. [Google Scholar] [CrossRef] [PubMed]
- Doane, W.W. Completion of Meiosis in Uninseminated Eggs of Drosophila melanogaster. Science 1960, 132, 677–678. [Google Scholar] [CrossRef]
- Lewis, O.L.; Farr, C.L.; Kaguni, L.S. Drosophila melanogaster Mitochondrial DNA: Completion of the Nucleotide Sequence and Evolutionary Comparisons. Insect Mol. Biol. 1995, 4, 263–278. [Google Scholar] [CrossRef]
- Mackay, T.F.C.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M.; et al. The Drosophila melanogaster Genetic Reference Panel. Nature 2012, 482, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Geng, D.; Guo, Q.; Liu, W.; Shufen, L.I.; Gao, W.; Wang, Y.; Zhang, M.; Wang, Y.; Yanzhen, B.U.; et al. Genomic Landscape of Mitochondrial DNA Insertions in 23 Bat Genomes: Characteristics, Loci, Phylogeny, and Polymorphism. Integr. Zool. 2021; Online ahead of print. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Lupotto, M.; Pellanda, P.; Santagostino, M.; Vitelli, V.; Giulotto, E. Mitochondrial DNA Insertions in the Nuclear Horse Genome. Anim. Genet. 2010, 41, 176–185. [Google Scholar] [CrossRef]
- Schiavo, G.; Hoffmann, O.I.; Ribani, A.; Utzeri, V.J.; Ghionda, M.C.; Bertolini, F.; Geraci, C.; Bovo, S.; Fontanesi, L. A Genomic Landscape of Mitochondrial DNA Insertions in the Pig Nuclear Genome Provides Evolutionary Signatures of Interspecies Admixture. DNA Res. 2017, 24, 487–498. [Google Scholar] [CrossRef]
- Lang, M.; Sazzini, M.; Calabrese, F.M.; Simone, D.; Boattini, A.; Romeo, G.; Luiselli, D.; Attimonelli, M.; Gasparre, G. Polymorphic NumtS Trace Human Population Relationships. Hum. Genet. 2012, 131, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Dayama, G.; Emery, S.B.; Kidd, J.M.; Mills, R.E. The Genomic Landscape of Polymorphic Human Nuclear Mitochondrial Insertions. Nucleic Acids Res. 2014, 42, 12640–12649. [Google Scholar] [CrossRef] [Green Version]
- Salas, A.; Schönherr, S.; Bandelt, H.J.; Gómez-Carballa, A.; Weissensteiner, H. Extraordinary Claims Require Extraordinary Evidence in Asserted MtDNA Biparental Inheritance. Forensic Sci. Int. Genet. 2020, 47, 102274. [Google Scholar] [CrossRef]
- Bai, R.; Cui, H.; Devaney, J.M.; Allis, K.M.; Balog, A.M.; Liu, X.; Schnur, R.E.; Shapiro, F.L.; Brautbar, A.; Estrada-Veras, J.I.; et al. Interference of Nuclear Mitochondrial DNA Segments in Mitochondrial DNA Testing Resembles Biparental Transmission of Mitochondrial DNA in Humans. Genet. Med. 2021, 23, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Pagnamenta, A.T.; Gleadall, N.; Sanchis-Juan, A.; Stephens, J.; Broxholme, J.; Tuna, S.; Odhams, C.A.; Ambrose, J.C.; Baple, E.L.; et al. Nuclear-Mitochondrial DNA Segments Resemble Paternally Inherited Mitochondrial DNA in Humans. Nat. Commun. 2020, 11, 1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
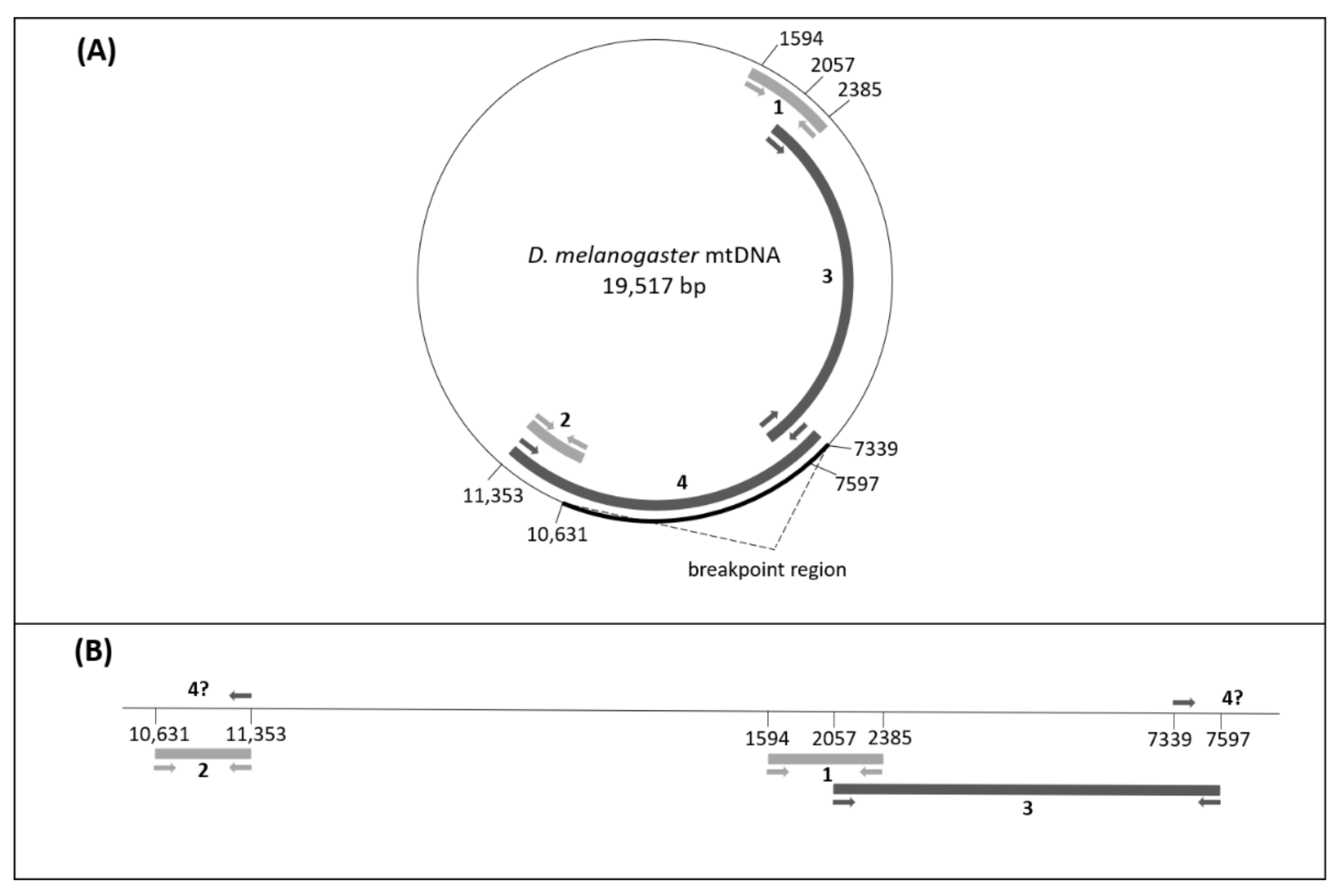
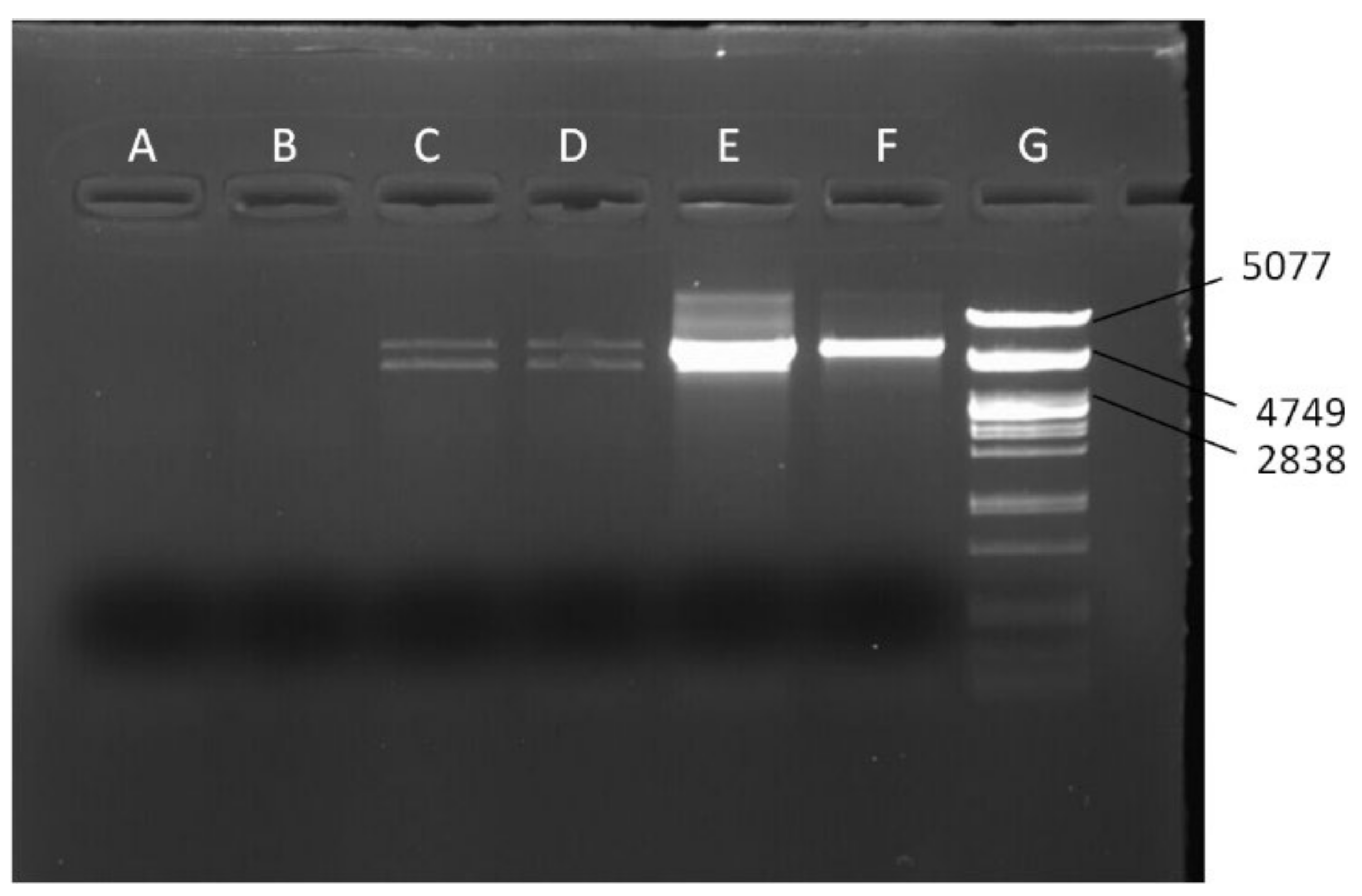
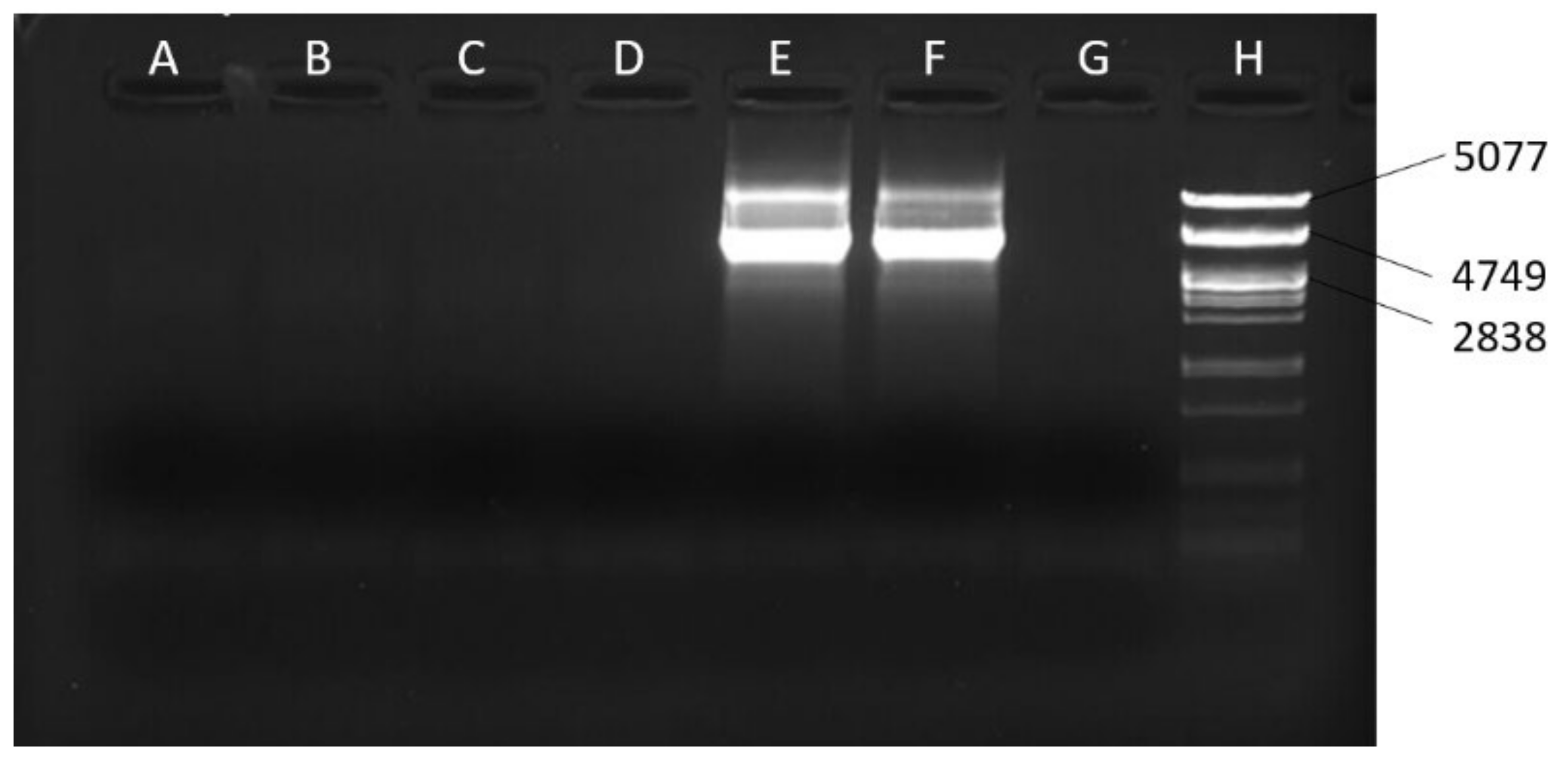
| Lines | Mitotype |
|---|---|
| mel;DGRP-820 | mel |
| siI;820 | siI |
| sm21;820 | siII |
| siI;w1118;+/+;+/+ | siI |
| sm21;w1118;+/+;+/+ | siII |
| mau12;w1118;+/+;+/+ | siIII |
| D. melanogaster Oregon R | mel |
| mel;y v f malbz/ y v f malbz; +/+;+/+ | mel |
| mel;FM6 B1/+;+/+;+/+ | mel |
| siI;w1118;CyO/Sp;TM6B/Dr | siI |
| siII;w1118;CyO/Sp;TM6B/Dr | siII |
| Reference | Primers | Sequence (5′ -> 3′) | Specificity | PCR Product Size (bp) | Tm (°C) |
|---|---|---|---|---|---|
| [30] | mel1594F | GCTGAATTAGGACATCCTGGAGC | mel against siI, siII, siIII | 791 | 61 |
| mel2385R | TCGAGTATCTACATCTATTCCAACG | ||||
| Present study | mel10631F | CGAAATTCCCATCCTC | mel against siI, siII, siIII | 722 | 52 |
| mel11353R | TTATCAGGGTCTCCCA | ||||
| [30] | siI_1737F | TCCTGATATAGCATTTCCA | siI against mel, siIII | 794 | 58 |
| siI_2531R | GTTAATCCTCCTACTGTG | ||||
| [31] | siII_1737F | CCCTGATATAGCATTCCCG | siII against mel, siIII | 794 | 58 |
| siII_2531R | GTTAACCCCCCTACTGTA | ||||
| Present study | 1588+ | GAATTAGGACATCCTGGAGCAT | siIII against mel, siI, siII | 786 | 61 |
| 2374− | GAGTATCAACGTCTATTCCAACTGTG | ||||
| Kindly provided by Maria Monastirioti | 1002(F) | TCGGAATAAGTTGAAGGATG | X chromosome against mtDNA | 767 | 52 |
| 2653(R) | TGCCATCCTGACTGCTCAGC | ||||
| Present study | 7339+ | AAGCATGAGTTAATAAATGAAA | mtDNA universal primers | 258 | 54 |
| 7597− | CCGTTTCTGCTTTAGTTC | ||||
| Present study | 7339+ | AAGCATGAGTTAATAAATGAAA | Used for the long PCR assay | 4014 | 56 |
| mel11353R | TTATCAGGGTCTCCCA | ||||
| Present study | mel2057F | TATTATTATCACTTCCAGTAC | Used with 7597− for the long PCR assay | 5540 | 54 |
| 7597− | CCGTTTCTGCTTTAGTTC |
| Line | Genotype |
|---|---|
| Stable line 1 | |
| Stable line 2 | |
| Stable line 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parakatselaki, M.-E.; Zhu, C.-T.; Rand, D.; Ladoukakis, E.D. NUMTs Can Imitate Biparental Transmission of mtDNA—A Case in Drosophila melanogaster. Genes 2022, 13, 1023. https://doi.org/10.3390/genes13061023
Parakatselaki M-E, Zhu C-T, Rand D, Ladoukakis ED. NUMTs Can Imitate Biparental Transmission of mtDNA—A Case in Drosophila melanogaster. Genes. 2022; 13(6):1023. https://doi.org/10.3390/genes13061023
Chicago/Turabian StyleParakatselaki, Maria-Eleni, Chen-Tseh Zhu, David Rand, and Emmanuel D. Ladoukakis. 2022. "NUMTs Can Imitate Biparental Transmission of mtDNA—A Case in Drosophila melanogaster" Genes 13, no. 6: 1023. https://doi.org/10.3390/genes13061023
APA StyleParakatselaki, M.-E., Zhu, C.-T., Rand, D., & Ladoukakis, E. D. (2022). NUMTs Can Imitate Biparental Transmission of mtDNA—A Case in Drosophila melanogaster. Genes, 13(6), 1023. https://doi.org/10.3390/genes13061023






