Abstract
Cynipoidea is a medium-sized superfamily of Hymenoptera with diverse lifestyles. In this study, 16 mitochondrial genomes were newly sequenced, 11 of which were the first obtained mitochondrial genomes in the family Liopteridae and four subfamilies (Anacharitinae, Aspicerinae, Figitinae, and Parnipinae) of Figitidae. All of the newly sequenced mitogenomes have unique rearrangement types within Cynipoidea, whereas some gene patterns are conserved in several groups. nad5-nad4-nad4L-nad6-cytb was remotely inverted and two rRNA genes were translocated to nad3 downstream in Ibaliidae and three subfamilies (Anacharitinae, Eucoilinae, and Parnipinae within Figitidae); two rRNA genes in Aspicerinae, Figitinae, and Liopteridae were remotely inverted to the cytb-nad1 junction; rrnL-rrnS was translocated to the cytb-nad1 junction in Cynipidae. Phylogenetic inference suggested that Figitidae was a polyphyletic group, while the Ibaliidae nested deep within Cynipoidea and was a sister-group to the Figitidae. These results will improve our understanding of the gene rearrangement of the mitogenomes and the phylogenetic relationships in the Cynipoidea.
1. Introduction
Cynipoidea is a medium-sized superfamily of Hymenoptera, including around 223 genera with 3200 species described worldwide [1,2]. It includes five generally accepted extant families, Austrocynipidae, Cynipidae, Figitidae, Ibaliidae, and Liopteridae [2,3]. Cynipoid wasps exhibit a wide range of lifestyles [1,4,5]. The most well-known members are phytophagous and easily observed as gall-formers, while the majority of the species are small parasitoids or hyperparasitoids. Although previous molecular and/or morphological studies contributed to elucidating the family or subfamily relationships within Cynipoidea [1,6,7], the phylogenetic relationships within the Cynipoidea are still unclear and need further study, especially the phylogenetic relationships within the family Figitidae and Cynipidae.
The typical insect mitochondrial genome is a circular molecule that is 14–19 kb in size and encodes 37 genes, including 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes [8]. Mitogenomes have been widely used for phylogenetics, though with the limitation of a relatively high evolutionary rate and the presence of base composition bias [8,9,10]. Most mitogenomes of insects are highly conserved, possessing the ancestral mitogenome arrangement; however, hymenopteran mitogenomes show extremely high rates of genome rearrangements [11,12,13,14]. tRNA rearrangements are widespread in Hymenoptera, with at least one tRNA rearrangement found in every sequenced hymenopteran species. Gene rearrangements are usually confined to specific lineages, which can help with phylogenetic reconstruction at lower taxonomic levels, such as the subfamily level in Braconidae [13]. At present, the complete or partial mitogenomes of only seven cynipoid species are available in GenBank (https://www.ncbi.nlm.nih.gov/; accessed on 20 January 2022). The synapomorphic mitochondrial gene rearrangement characters and their phylogenetic utility could not be fully assessed due to limited taxon sampling in the early studies.
In this study, 16 mitogenomes of Cynipoidea were newly sequenced by next generation sequencing (NGS), and 11 of them were the first obtained mitogenomes in the family Liopteridae and four subfamilies (Anacharitinae, Aspicerinae, Figitinae, and Parnipinae) of Figitidae. The obtained information from the study will also facilitate future phylogenetic research of Cynipoidea. Furthermore, we analyzed the main features of the newly generated mitogenomes and those of other Cynipoidea species. We also analyzed gene rearrangement patterns. Finally, the phylogeny of Cynipoidea was reconstructed by combining the available mitogenomes.
2. Materials and Methods
2.1. Sample Identification and DNA Extraction
All 16 newly sequenced samples were identified based on the morphology of adults according to the taxonomic literature (Table S1). All specimens were initially preserved in 100% ethanol and then stored at 4 °C before DNA extraction. Whole genomic DNA was non-destructively extracted from every sample using the DNeasy tissue kit (Qiagen, Hilden, Germany), modified from previous studies [15,16]. Voucher specimens were deposited in the Institute of Insect Sciences, Zhejiang University (Voucher specimen numbers: ZJUH_20220001- ZJUH_20220016, Table S1).
2.2. Next-Generation Sequencing and Assembly
All libraries were constructed using the VAHTS® Universal DNA Library Prep Kit. Whole-genome data were generated on the Illumina NovaSeq platform (Illumina, San Diego, CA, USA) with a PE150 strategy (2 × 150 base, paired-end reads).
More than 2 GB of raw data of each sample was obtained. The raw reads were checked by FastQC v0.11.9 [17], with adapter contamination trimmed by Trimmomatic [18]. The target mitochondrial reads were filtered out using BLAST v2.9.0+ (BLASTn, E-value cutoff 1 × 10−5) against a reference dataset of published Cynipoidea mitogenomes [19]. The mitochondrial reads were assembled by SPAdes v3.0 [20] and IDBA v1.1.3 [21] with default parameters, respectively. Two assemblies were then integrated with GENEIOUS v2020.0.5 (Biomatters Ltd., San Diego, CA, USA).
2.3. Mitochondrial Genome Annotation and Analysis
Assembled contigs were initially annotated using the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py; accessed on 15 June 2021) [22]. The start and stop positions of 13 PCGs were adjusted manually and corrected by aligning published data of Cynipoidea species in GenBank. The putative tRNA genes were confirmed by the tRNAscan-SE search server with their homologs from related species [23]. The obtained mitogenomes were submitted to GenBank (Accession number: OM677820-OM677835, Table 1).

Table 1.
Information of mitochondrial genomes used in phylogenetic analysis.
The nucleotide composition of all components and the relative synonymous codon usage (RSCU) of PCGs were estimated using MEGA 11.0 [24]. The base composition values (AT and GC-skews) were calculated using the following formulas: AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) [25]. The numbers of the synonymous substitutions (Ks) and non-synonymous substitutions (Ka), and the ratios of Ka/Ks for each PCG were calculated in the DnaSP 6.0 [26]. The gene rearrangement of all protein-coding genes, all tRNAs, and two rRNAs in the 20 cynipoid mitogenomes were analyzed by comparison with the ancestral mitogenomes and with each other.
2.4. Phylogenetic Analysis
A total of 20 mitogenomes representing the superfamily Cynipoidea, including 16 newly obtained taxa, were used for phylogenetic analyses. Two species from Platygastridae, Platygaster sp. and Trissolcus basalis were used as outgroups (Table 1). The PCGs were realigned using the G-INS-i algorithm implemented in MAFFT v7.464 [27]. Bayesian inference analysis (BI) was conducted with MrBayes v3.2.7a [28] using the most optimal partition schemes and best model schemes (Table S2) acquired by PartitionFinder v1.1.1 [29]. Four independent Markov chains were run for 100 million generations, with tree sampling occurring every 1000 generations and a burn-in of 25% of the trees. The stationarity of the run was assessed by Tracer v1.7. (ESS values > 200) [30]. Maximum likelihood (ML) analysis was performed with RAxML-HPC2 v8.2.12 [31] under the GTRGAMMA model. A total of 200 runs for different individual partitions were conducted with 1000 bootstrap replicates.
3. Results and Discussion
3.1. General Features of Mitochondrial Genomes
We obtained 16 new mitogenomes from the taxa of Cynipoidea. The 37 typical genes were identified in each of the newly sequenced mitogenomes except for Trybliographa sp. (missing trnF), Figites sp. 1 (missing trnW, trnN, trnL1, trnS2, and trnV), Figites sp. 2 (missing trnL1), and Pujadella villari (missing trnQ) (Figure 1). The complete control region (CR) of all the species failed to be assembled, possibly due to low similarity between reference and high A and T contents, common in insect mitogenomes, especially in Hymenoptera [19].
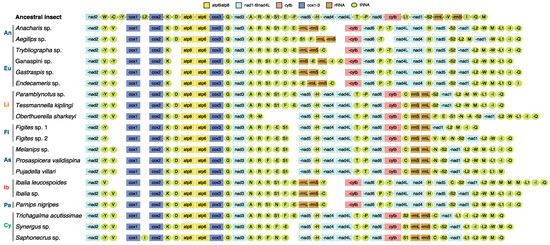
Figure 1.
Mitogenomic architecture of the Cynipoidea referenced with the ancestral insect mitochondrial genome. An, Anacharitinae; Eu, Eucoilinae; Li, Liopteridae; Fi, Figitinae; As, Aspicerinae; Ib, Ibaliidae; Pa, Parnipinae; Cy, Cynipidae. Families are shown in different colors.
The A + T content for the sequenced region of the mitogenomes in the Cynipoidea ranged from 79.36% (Trybliographa sp.) to 87.01% (Paramblynotus sp.) (Table 2). There was no significant difference in base composition among the species of Cynipoidea. Relative high A + T content in the mitogenomes is not unusual in Hymenoptera compared with other orders [32].

Table 2.
Base composition of 16 mitochondrial genomes in Chalcidoidea.
3.2. Base Composition, Codon Usage, and Evolutionary Rate
All 13 PCGs were identified in the newly generated mitogenomes, with sizes ranging from 10,986 bp (Figites sp. 1) to 11,381 bp (Anacharis sp.). The entire A + T content of all the PCGs ranged from 76.81% (Pu. villari) to 85.66% (Ibalia sp.) (Table 2). The AT-skew in all Cynipoidea mitogenomes was negative. The GC-skew in Anacharitinae, Eucoilinae, and Liopteridae was also negative, whereas Paramblynotus sp. in Lio-pteridae was positive (Table 2), which is an unusual feature of cynipoid mitogenomes. Two rRNA genes (rrnS and rrnL) were identified in all mitogenomes. The length of rrnS ranged from 792 bp (Aegilips sp.) to 879 bp (Endecameris sp.), and the size of rrnL ranged from 1231 bp (Ganaspini sp.) to 1464 bp (Pu. villari) (Table S3).
The relative synonymous codon usage (RSCU) values of all four subfamilies were plotted in Figure 2, and all possible synonymous codons of the 22 amino acids were present. A or T nucleotides were used with higher frequency in the third codon position than other nucleotides, and A was used more often than T. The four most frequently used codons—AUA (Met), AUU (Ile), UUA (Leu2), and UUU (Phe)—were observed (Figure 2, Table S4). These results are consistent with published mitogenomes of other wasps [33,34].
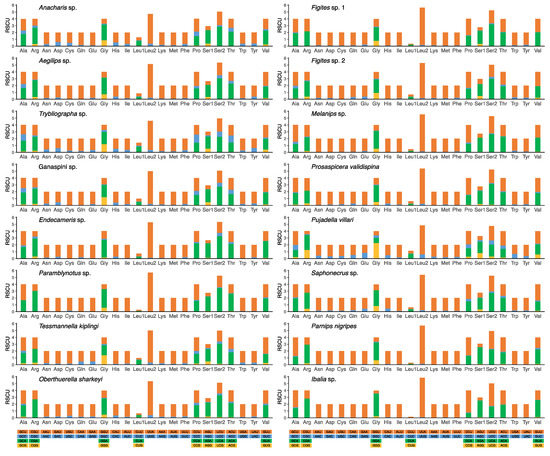
Figure 2.
Relative synonymous codon usage (RSCU) of 16 cynipoid mitogenomes. Codon families are provided on the X-axis along with the different combinations of synonymous codons that code for that amino acid. RSCU is defined on the Y-axis.
Ka (nonsynonymous substitutions) and Ks (synonymous substitutions) are used as indicators of selective pressure [35]. The evolutionary rates (Ka, Ks, and Ka/Ks) of PCGs vary considerably among genes (Figure 3, Table S5). In Cynipoidea, the Ka values of 13 PCGs ranged from 0.1177 (cox1) to 0.3506 (atp8), and Ks values ranged from 0.2566 (nad6) to 0.4494 (cox1). The average Ka/Ks ratios were estimated to investigate the evolutionary rates of Cynipoidea PCGs. Ratios ranged from 0.2619 (cox1) to 1.1850 (nad6). The genes nad2, atp8, and nad6 > 1, indicating that they evolved at a faster rate in Cynipoidea. This result was similar to that of the Apoidea [36] and Ichneumonidae [37] in Hymenoptera.
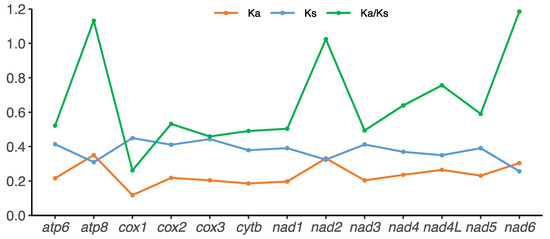
Figure 3.
Non-synonymous (Ka), synonymous substitutional (Ks) rates and the ratios of Ka/Ks of 13 protein coding genes (PCGs) in mitochondrial genomes of all sequenced species in Cynipoidea.
3.3. Gene Rearrangements
Compared with the putative ancestral mitogenome of insects, the mitogenomes of all the Cypoidea in this study are extremely variable, and all PCGs, tRNAs, and rRNAs had various degrees of rearrangement (Figure 2).
Gene nad2 was locally inverted in all Cynipoidea species, as previously observed in two Chalcidoidea species, Megaphragma amalphitanum and Eurytoma sp. [38]. In addition, a large block, “nad5-nad4-nad4L-nad6-cytb”, was remotely inverted in Ibaliidae and three subfamilies (Anacharitinae, Eucoilinae, and Parnipinae) within Figitidae, which is a unique gene rearrangement pattern in Cynipoidea and had not been observed in other Hymenoptera mitogenomes.
Two rRNA genes were translocated to nad3 downstream in Ibaliidae and three subfamilies (Anacharitinae, Eucoilinae, and Parnipinae) within Figitidae, which is consistent with the large block PCGs rearrangement mentioned above. In addition, the two rRNA genes in Aspicerinae, Figitinae, and Liopteridae was remotely inverted to the cytb-nad1 junction. In Cynipidae, rrnL-rrnS was translocated to the cytb-nad1 junction.
The tRNA genes exhibited an extremely diverse rearrangement in Cynipoidea. Except for the stable positions of three tRNA genes (trnK, trnG, and trnH), the other tRNA genes were more or less rearranged. The hot spots of rearrangement were concentrated in two tRNA clusters, trnI-trnQ-trnM and trnA-trnR-trnN-trnS1-trnE-trnF. The two tRNA clusters were rearranged to varying degrees in all species. Patterns of tRNA rearrangement were conserved in several groups. trnD was relatively stable and only rearranged in Eucoilinae. In Figitinae and Aspicerinae, the tRNA cluster trnA-trnR-trnN-trnS1-trnE-trnF was shuffled into trnA-trnR-trnF-trnE-trnS1. In Cynipidae, trnL1 was translocated from upstream to downstream of nad1, and the cluster trnL2-trnW-trnM-trnQ was upstream of nad2. In Liopteridae, Ibaliidae, and two subfamilies (Anacharitinae and Parnipinae) of Figitidae, the cluster trnL1-trnI-trnQ was translocated upstream of nad2.
In conclusion, our analyses indicate that the rearrangement types of all newly sequenced mitogenomes in this study are novel within Cynipoidea. The gene rearrangements in Cynipoidea are randomly distributed, although they are conserved in several groups.
3.4. Phylogenetic Analyses
Concerning the phylogenetic relationships of Cynipoidea, all analyses based on the PCGs matrix and two inference methods (BI and ML) generated congruent results with high nodal supports (Figure 4). The results indicate that the Figitidae is a polyphyletic group, consistent with the results based on UCEs [4]. The family Cynipidae is recovered as monophyletic by our data, as most previous studies assumed [1], but this contradicts the result of Blaimer et al. [4]. Adding more members to this family may help to show whether it is monophyletic or not. Ibaliidae was formerly thought to be an early-branching cynipoid in most studies [6,7]. However, Blaimer et al. proposed that Ibaliidae nested far inside Cynipoidea and was a sister-group to Figitidae [4]. Our research likewise came up with a similar result.
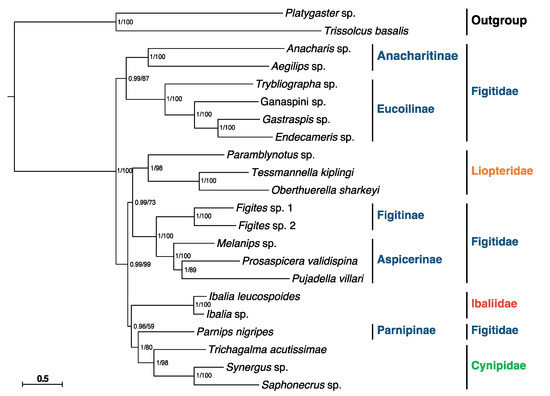
Figure 4.
Phylogenetic analyses of Cynipoidea based on nucleotide datasets of 13 PCGs. The scale bar corresponds to the estimated number of substitutions per site. Numbers separated by a slash on the node are posterior probability (PP) and bootstrap value (BV).
4. Conclusions
In this study, 16 Cynipoidea mitogenomes were newly obtained using the next-generation sequencing method, in which the mitogenomes of the family Liopteridae and four subfamilies Anacharitinae, Aspicerinae, Figitinae, and Parnipinae of Figitidae were first reported. The mitogenomes of all the Cypoidea in this research are highly variable. All of the newly sequenced mitogenomes in this study have unique rearrangement types within Cynipoidea. The gene rearrangements in Cynipoidea are randomly distributed, though there are gene patterns conserved in several groups, for instance, nad5-nad4-nad4L-nad6-cytb was remotely inverted and two rRNA genes were translocated to nad3 downstream in Ibaliidae, and three subfamilies Anacharitinae, Eucoilinae, and Parnipinae within Figitidae; two rRNA genes in Aspicerinae, Figitinae, and Liopteridae were remotely inverted to the cytb-nad1 junction; rrnL-rrnS was translocated to the cytb-nad1 junction in Cynipidae. The BI and ML analysis showed consistent topology and indicated that Figitidae was a polyphyletic group and Ibaliidae nested far inside Cynipoidea and was a sister-group to Figitidae. Nevertheless, these results provide valuable information for understanding the evolution of Cynipoidea. Denser taxon sampling provides more accurate and comprehensive information for the further analysis of gene arrangement and the evolutionary history of Cynipoidea.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13050914/s1, Table S1: Collection information of Cynipoidea species in this study; Table S2: The best schemes of partition and substation models selected in nucleotide dataset; Table S3: Length of tRNA and rRNA genes in the Cynipoidea mitogenomes; Table S4: Relative synonymous codon usage in 16 Cynipoidea mitogenomes; Table S5: Synonymous and nonsynonymous substitutional analysis of 13 protein coding genes.
Author Contributions
Conceptualization, supervision and funding acquisition, X.C. and P.T.; investigation, X.S., Z.L. and R.Y.; writing—review and editing, X.S., X.C. and P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Guangdong Laboratory of Lingnan Modern Agriculture Project (NT2021003), the Key International Joint Research Program of National Natural Science Foundation of China (31920103005), the General Program of National Natural Science Foundation of China (32070467) and the Key R&D Program of Zhejiang Province (2020C02003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The new mitogenome assemblies and annotation data in this study have been submitted to the GenBank database under accession numbers: OM677820-OM677835.
Acknowledgments
We thank Cornelis van Achterberg for linguistic assistance and Xiqian Ye for providing access to the cluster computer system used for the phylogenetic analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ronquist, F. Phylogeny, classification and evolution of the Cynipoidea. Zool. Scr. 1999, 28, 139–164. [Google Scholar] [CrossRef] [Green Version]
- Huber, J.T. Biodiversity of Hymenoptera. In Insect Biodiversity: Science and Society, 2nd ed.; Foottit, R.G., Adler, P.H., Eds.; Wiley-Blackwell: Oxford, UK, 2017; pp. 419–461. [Google Scholar]
- Buffington, M.L.; Forshage, M.; Liljeblad, J.; Tang, C.T.; van Noort, S. World Cynipoidea (Hymenoptera): A key to higher-level groups. Insect Syst. Divers. 2020, 4, 1. [Google Scholar] [CrossRef]
- Blaimer, B.B.; Gotzek, D.; Brady, S.G.; Buffington, M.L. Comprehensive phylogenomic analyses re-write the evolution of parasitism within cynipoid wasps. BMC Evol. Biol. 2020, 20, 155. [Google Scholar] [CrossRef]
- Egan, S.P.; Hood, G.R.; Martinson, E.O.; Ott, J.R. Cynipid gall wasps. Curr. Biol. 2018, 28, R1370–R1374. [Google Scholar] [CrossRef] [Green Version]
- Buffington, M.L.; Nylander, J.A.A.; Heraty, J.M. The phylogeny and evolution of Figitidae (Hymenoptera: Cynipoidea). Cladistics 2007, 23, 403–431. [Google Scholar] [CrossRef]
- Ronquist, F.; Nieves-Aldrey, J.-L.; Buffington, M.L.; Liu, Z.; Liljeblad, J.; Nylander, J.A.A. Phylogeny, evolution and classification of gall wasps: The plot thickens. PLoS ONE 2015, 10, e0123301. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [Green Version]
- Dowton, M.; Castro, L.R.; Austin, A.D. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebr. Syst. 2002, 16, 345–356. [Google Scholar] [CrossRef]
- Castro, L.R.; Dowton, M. Mitochondrial genomes in the Hymenoptera and their utility as phylogenetic markers. Syst. Entomol. 2007, 32, 60–69. [Google Scholar] [CrossRef]
- Dowton, M.; Austin, A.D. Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the hymenoptera. Mol. Biol. Evol. 1999, 16, 298–309. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; van Achterberg, C.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genom. 2010, 11, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Wei, S.J.; Tang, P.; Wu, Q.; Shi, M.; Sharkey, M.J.; Chen, X.X. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol. Evol. 2016, 8, 2651–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.Y.; Cao, L.J.; Chen, P.Y.; Chen, X.X.; van Achterberg, K.; Hoffmann, A.A.; Liu, J.X.; Wei, S.J. Comparative mitogenomics and phylogenetics of the stinging wasps (Hymenoptera: Aculeata). Mol. Phylogenet. Evol. 2021, 159, 107119. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.T.P.; Moore, W.; Melchior, L.; Worobey, M. DNA extraction from dry museum beetles without conferring external morphological damage. PLoS ONE 2007, 2, e272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzold, F.; Zilli, A.; Hundsdoerfer, A.K. Advantages of an easy-to-use DNA extraction method for minimal-destructive analysis of collection specimens. PLoS ONE 2020, 15, e0235222. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC. Available online: https://qubeshub.org/resources/fastqc (accessed on 8 January 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Tang, P.; Zhu, J.C.; Zheng, B.Y.; Wei, S.J.; Sharkey, M.; Chen, X.X.; Vogler, A.P. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenet. Evol. 2019, 131, 8–18. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Son, P.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Bernt, M.; Donath, A.; Juehling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Puetz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Carlos Sanchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Kruitwagen, A.; Wertheim, B.; Beukeboom, L.W. Artificial selection for nonreproductive host killing in a native parasitoid on the invasive pest, Drosophila suzukii. Evol. Appl. 2021, 14, 1993–2011. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Oliveira, D.C.S.G.; Raychoudhury, R.; Lavrov, D.V.; Werren, J.H. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol. Biol. Evol. 2008, 25, 2167–2180. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.C.; Tang, P.; Zheng, B.Y.; Wu, Q.; Wei, S.J.; Chen, X.X. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int. J. Biol. Macromol. 2018, 118, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zheng, B.Y.; Zhu, J.C.; van Achterberg, C.; Tang, P.; Chen, X.X. The first two mitochondrial genomes of wood wasps (Hymenoptera: Symphyta): Novel gene rearrangements and higher-level phylogeny of the basal hymenopterans. Int. J. Biol. Macromol. 2019, 123, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, B.Y.; Cao, L.J.; Tang, P.; van Achterberg, K.; Hoffmann, A.A.; Chen, H.Y.; Chen, X.X.; Wei, S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenet. Evol. 2018, 124, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.Y.; Han, Y.Y.; Yuan, R.Z.; Liu, J.X.; Tang, P.; van Achterberg, C.; Chen, X.X. Mitochondrial genomes yield insights into the basal lineages of Ichneumonid wasps (Hymenoptera: Ichneumonidae). Genes 2022, 13, 218. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.C.; Fan, Q.; Tian, Y.; Wang, F.; Chen, X.X.; Werren, J.H.; Ye, G.Y. Mitochondrial DNA and their nuclear copies in the parasitic wasp Pteromalus puparum: A comparative analysis in Chalcidoidea. Int. J. Biol. Macromol. 2019, 121, 572–579. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).