Impact of Group II Baculovirus IAPs on Virus-Induced Apoptosis in Insect Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Viral Bacmid, Genes, Reagents, and Cells
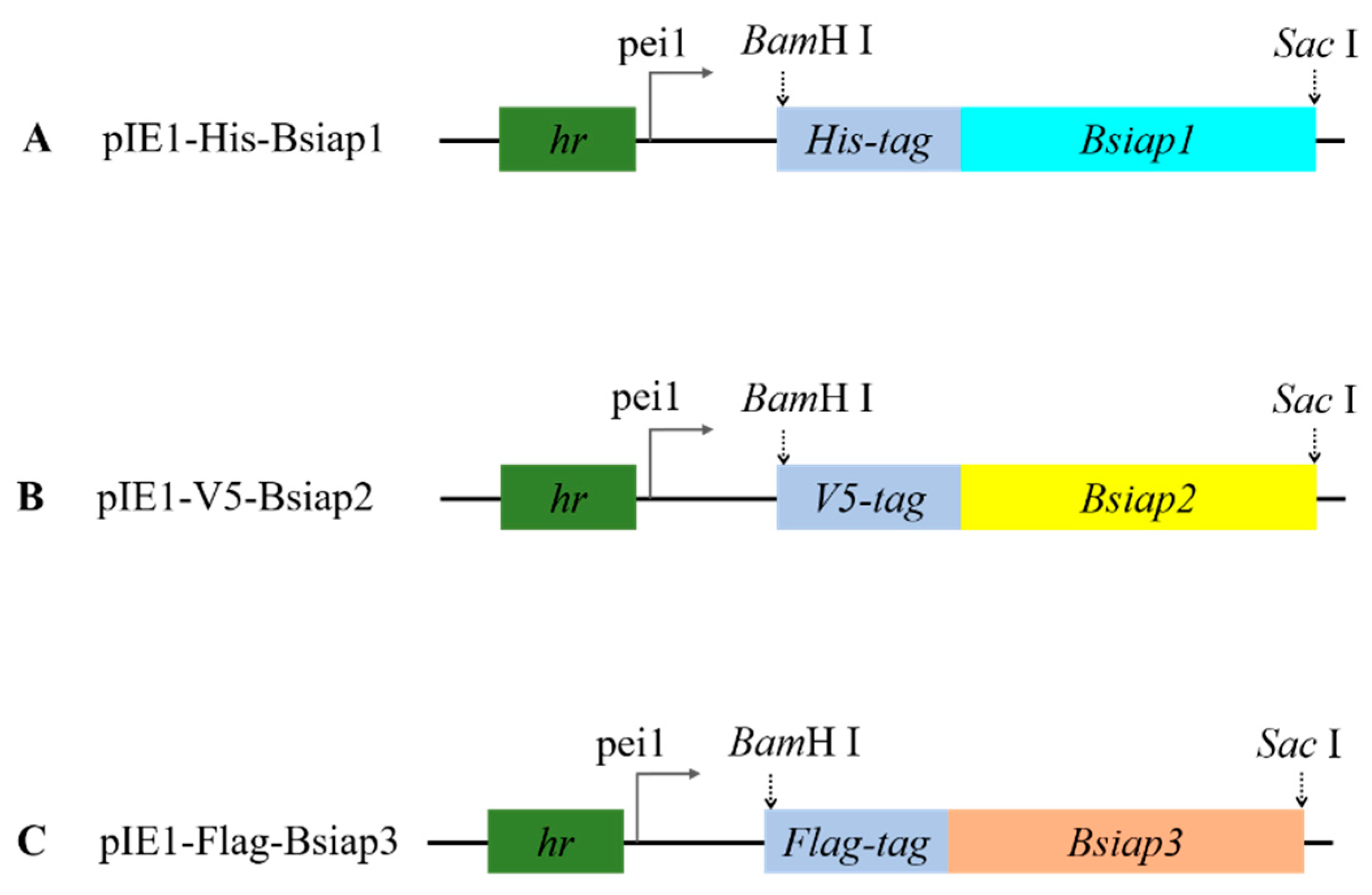
2.2. Construction of Transient Expression Vector
2.3. Transfection of Insect Cells
2.4. Quantitative PCR
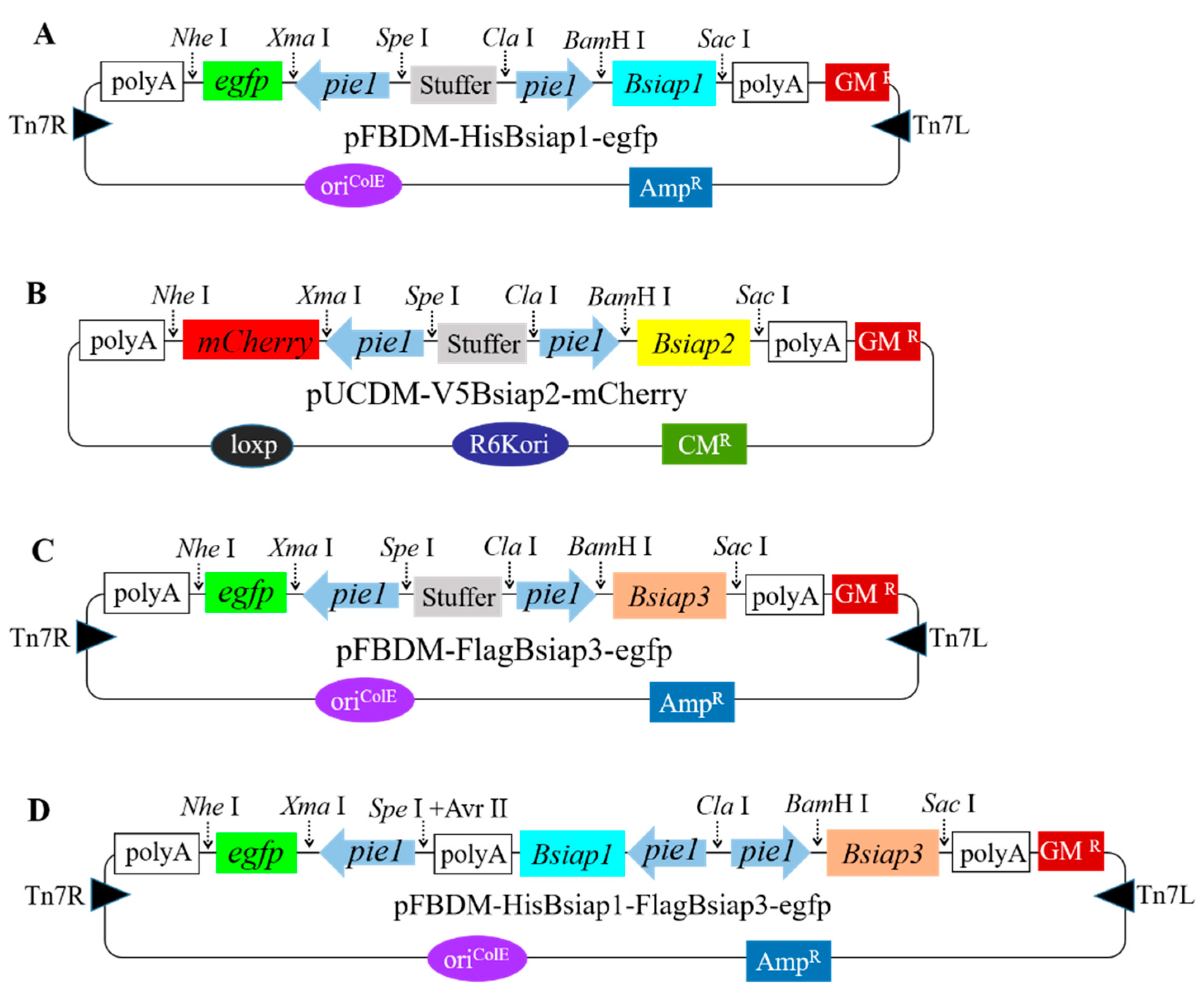
2.5. Construction of Baculovirus Donor Vectors Bearing Bsiaps Expression Cassettes
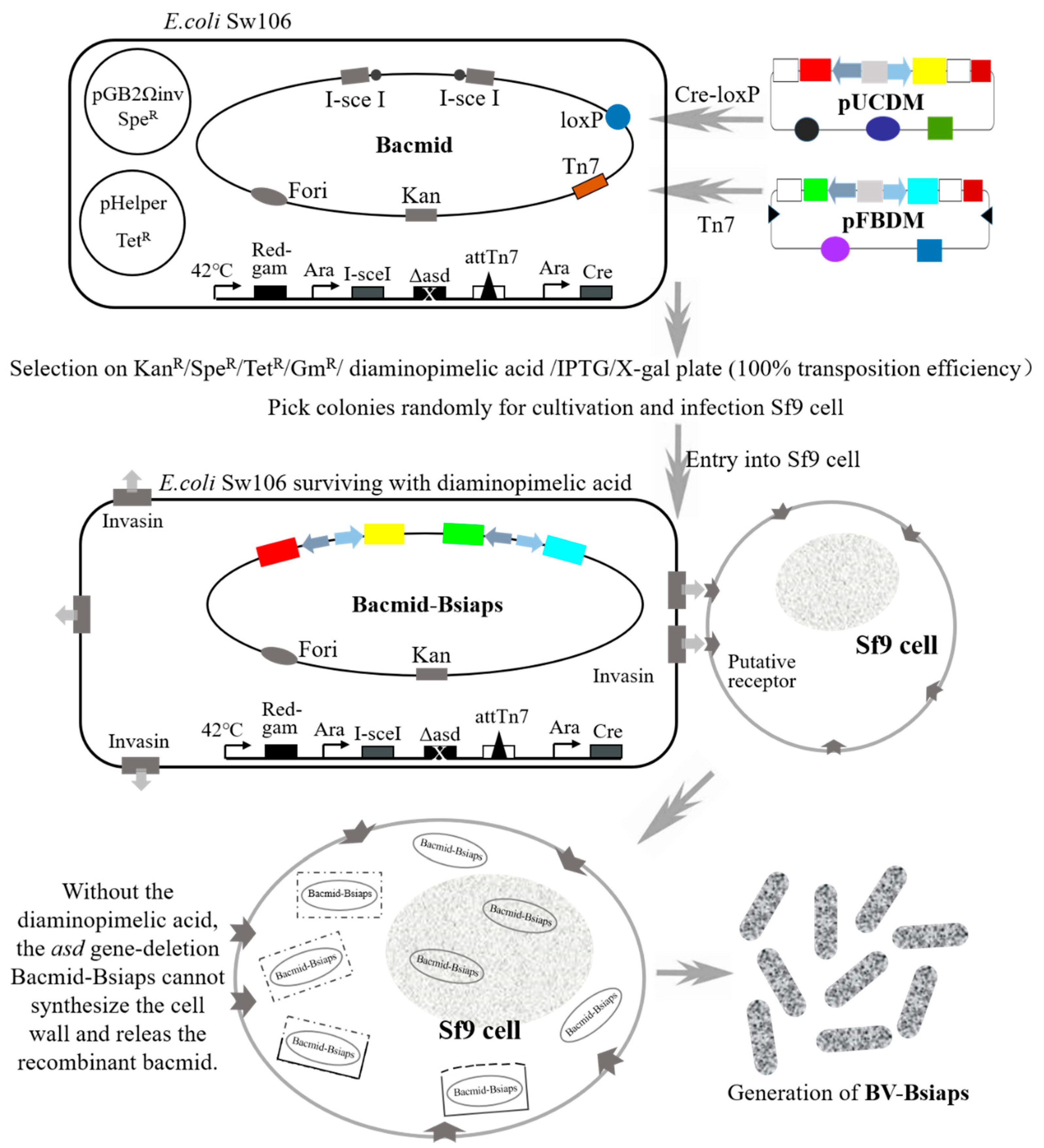
2.6. Introduction of Bsiaps into a Bacmid
2.7. Production of Recombinant Baculovirus
2.8. Western Blot Analysis
2.9. Determination of Recombinant Baculovirus Titer
2.10. Determination of Sf9 Apoptosis Rate by Flow Cytometry
2.11. Statistical Analysis
3. Results
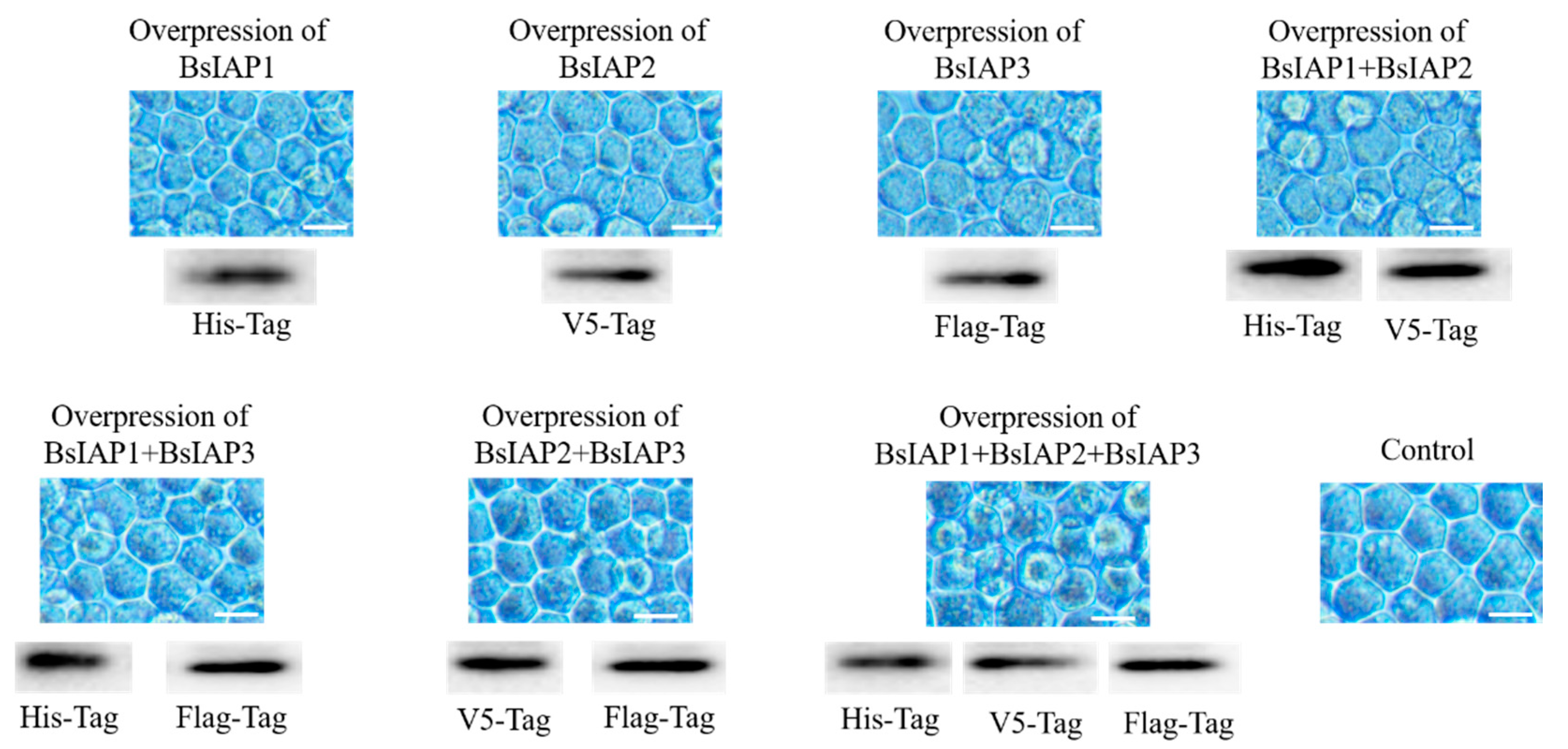
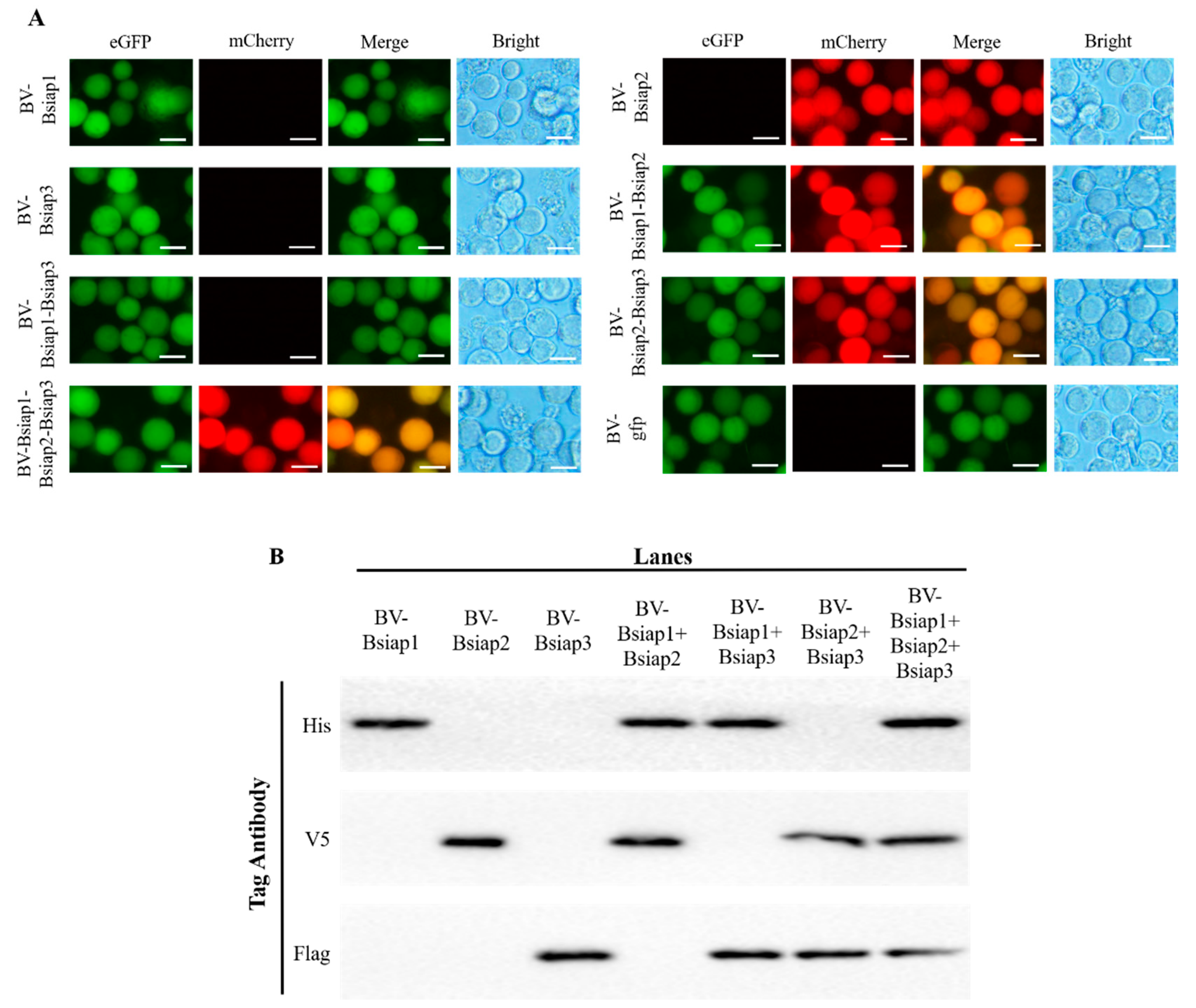
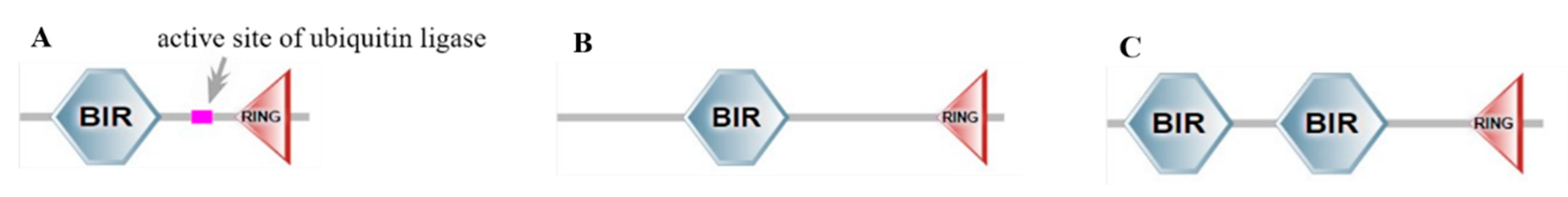
3.1. Transfection and Identification of BsIAPs
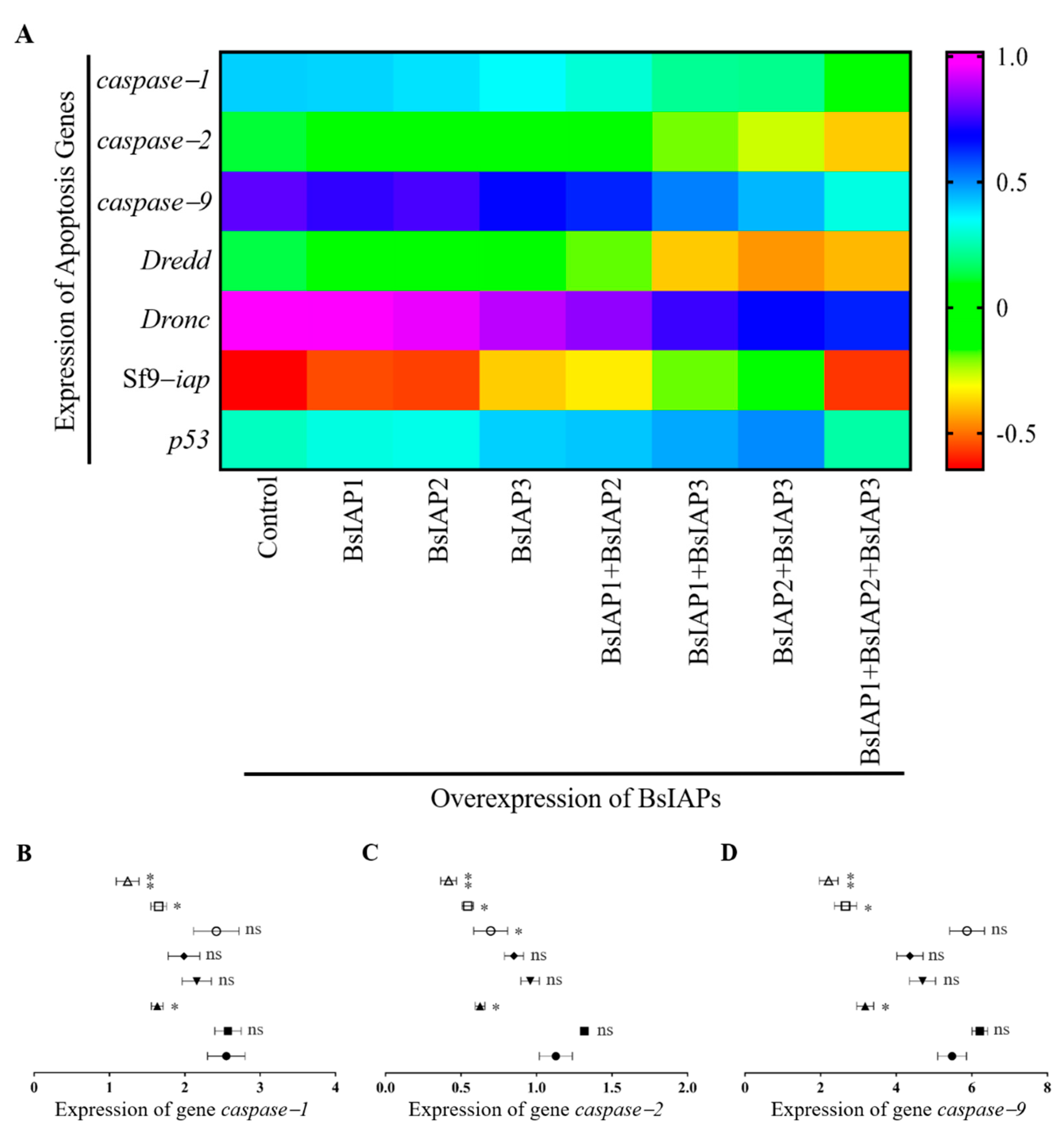
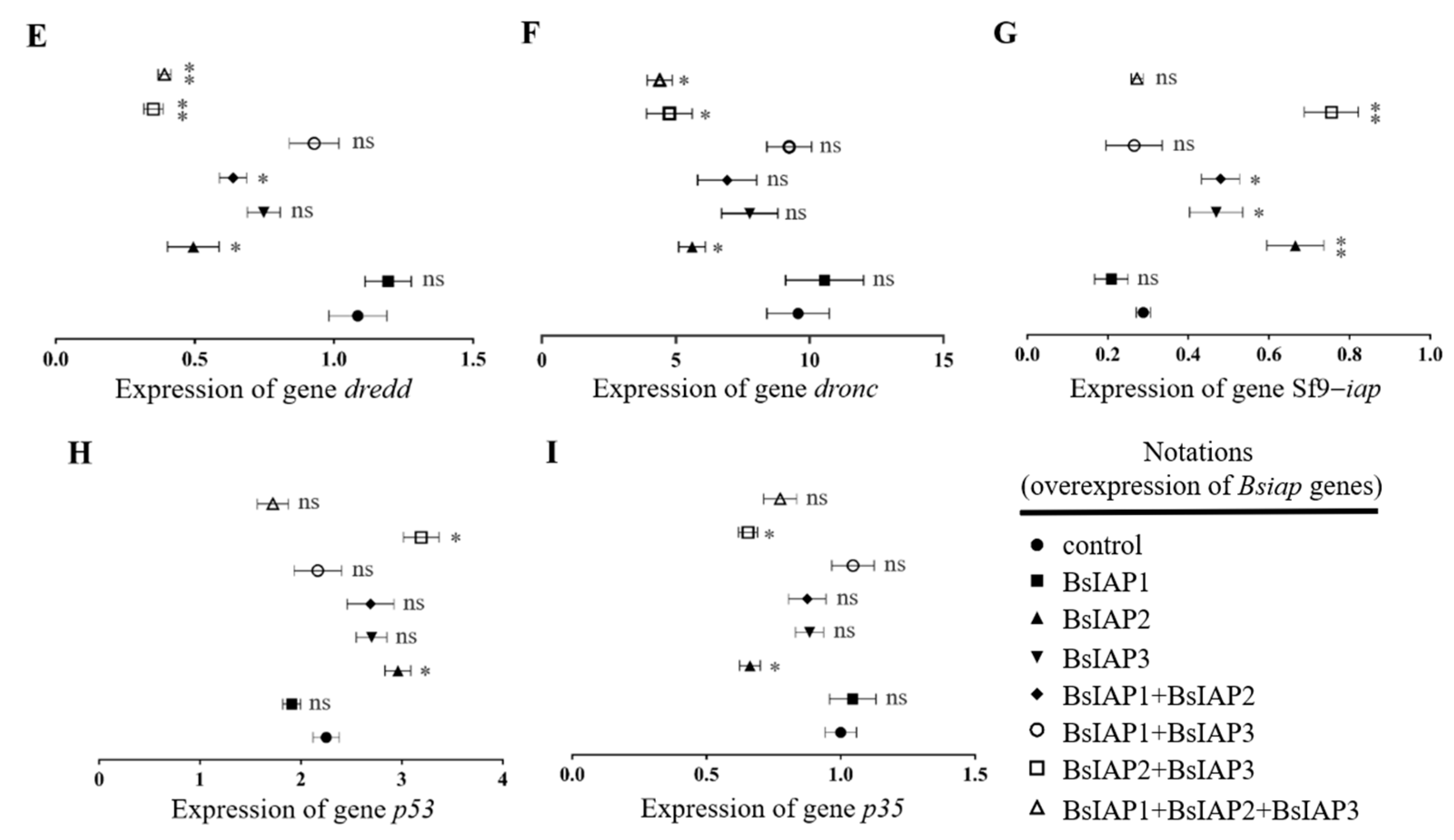
3.2. Influence of BsIAPs on Intracellular Apoptosis Gene Expression
3.3. Generation of Recombinant Baculovirus for Stable Expression of BsIAPs
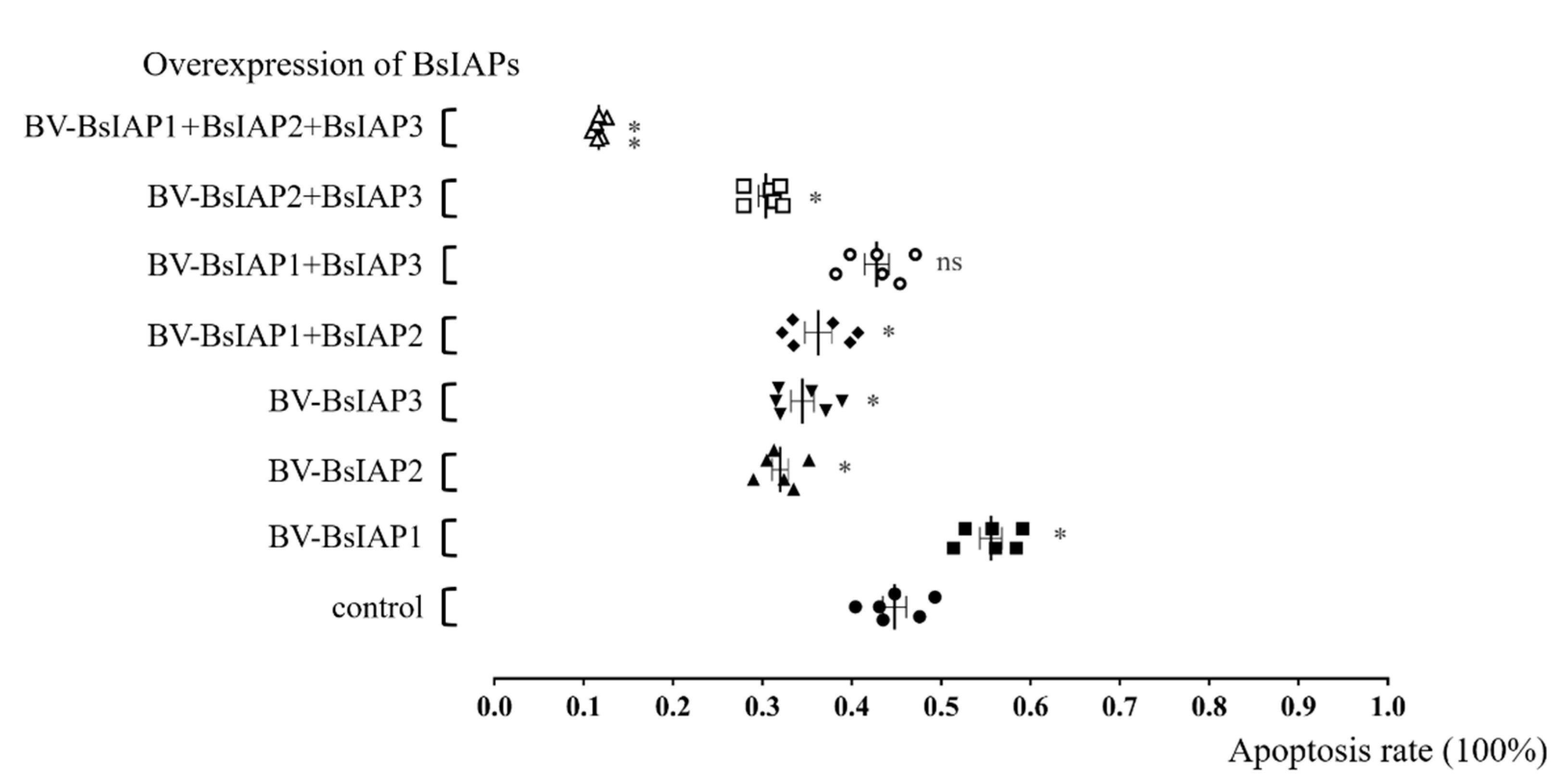
3.4. Influence of BsIAPs on Virus-Induced Apoptosis Rate of Sf9 Cells
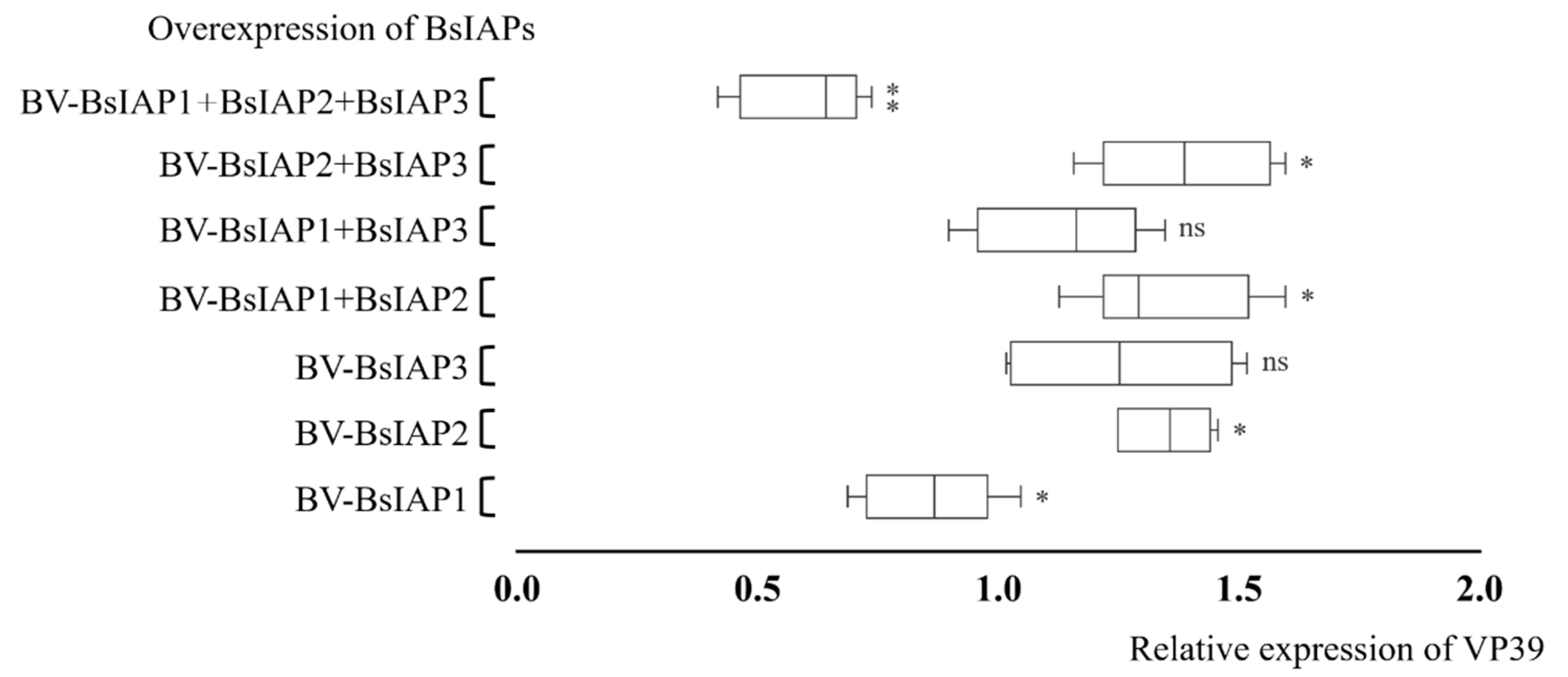
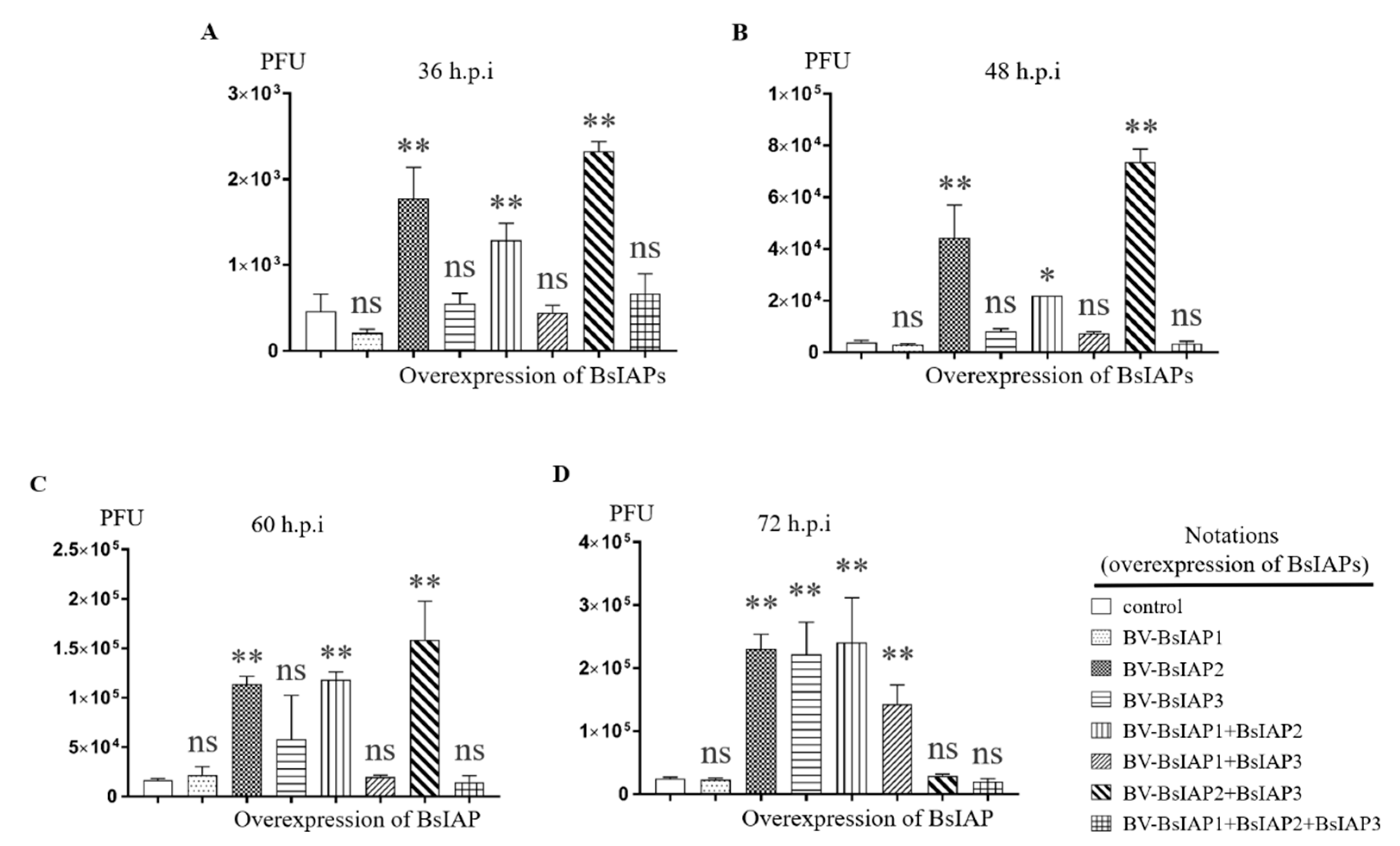
3.5. Effect of BsIAPs on Baculovirus Proliferation Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, H.; Wang, J.; Gao, H.; Li, S.; Zhang, Y.; Zheng, N. Heat-induced apoptosis and gene expression in bovine mammary epithelial cells. Anim. Prod. Sci. 2016, 56, 918. [Google Scholar] [CrossRef]
- Bedoui, S.; Herold, M.J.; Strasser, A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020, 21, 678–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Yu, X.F.; Qu, S.C.; Zhang, R.; Qu, X.R.; Chen, Y.P.; Ma, X.Y.; Sui, D.Y. Anti-proliferative effect of Juglone from Juglans mandshurica Maxim on human leukemia cell HL-60 by inducing apoptosis through the mitochondria-dependent pathway. Eur. J. Pharmacol. 2010, 645, 14–22. [Google Scholar] [CrossRef]
- Garrido, C.; Brunet, M.; Didelot, C.; Zermati, Y.; Schmitt, E.; Kroemer, G. Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties. Cell Cycle 2006, 5, 2592–2601. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Wang, H.; Guo, J.; Peng, M.; Ji, H.; Yang, H.; Liu, B.; Wang, J.; Zhang, X.; Li, S. Hsp70 suppresses apoptosis of BRL cells by regulating the expression of Bcl-2, cytochrome C, and caspase 8/3. Cell Dev. Biol. Anim. 2016, 52, 568–575. [Google Scholar] [CrossRef]
- Mace, P.D.; Shirley, S.; Day, C.L. Assembling the building blocks: Structure and function of inhibitor of apoptosis proteins. Cell Death Differ. 2010, 17, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Crook, N.E.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993, 67, 2168–2174. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Yanagimoto, K.; Kobayashi, M. Identification and functional analysis of Hyphantria cunea nucleopolyhedrovirus iap genes. Virol. N. Y. 2004, 321, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Hanako, I.; Hisanor, B.; Toru, S.; Susumu, K. The BIR and BIR-like domains of Bombyx mori nucleopolyhedrovirus IAP2 protein are required for efficient viral propagation. Biochem. Biophys. Res. Commun. 2014, 454, 581–587. [Google Scholar]
- Kasof, G.M.; Gomes, B.C. Livin, a Novel Inhibitor of Apoptosis Protein Family Member. J. Biol. Chem. 2001, 276, 3238–3246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Deveraux, Q.L.; Maeda, S.; Stennicke, H.R.; Hammock, B.D.; Reed, J.C. Cloning and characterization of an inhibitor of apoptosis protein (iap) from bombyx mori. BBA—Mol. Cell Res. 2001, 1499, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Luque, T.; Finch, R.; Crook, N.; O’Reilly, D.R.; Winstanley, D. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001, 82, 2531–2547. [Google Scholar] [CrossRef]
- Ikeda, M.; Yamada, H.; Ito, H.; Kobayashi, M. Baculovirus IAP1 induces caspase-dependent apoptosis in insect cells. J. Gen. Virol. 2011, 92, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Shibuya, M.; Kobayashi, M.; Ikeda, M. Baculovirus Lymantria dispar multiple nucleopolyhedrovirus iap2 and iap3 do not suppress apoptosis, but trigger apoptosis of insect cells in a transient expression assay. Virus Genes 2012, 45, 370. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lange, J.D.; Chen, X.; Oers, M.; Vlak, J.M.; Westenberg, M. Functional analysis of two inhibitor of apoptosis (iap) orthologs from Helicoverpa armigera nucleopolyhedrovirus. Virus Res. 2012, 165, 107–111. [Google Scholar] [CrossRef]
- Yan, F.; Deng, X.; Yan, J.; Wang, J.; Yao, L.; Lv, S. Functional analysis of the inhibitor of apoptosis genes in Antheraea pernyi nucleopolyhedrovirus. J. Microbiol. 2010, 48, 199–205. [Google Scholar] [CrossRef]
- Long, S.; Wilson, M.; Bengtén, E.; Clem, L.W.; Miller, N.W.; Chinchar, V.G. Identification and characterization of a FasL-like protein and cDNAs encoding the channel catfish death-inducing signaling complex. Immuno-Genetics 2004, 56, 518. [Google Scholar] [CrossRef] [Green Version]
- Zanotto, P.; Kessing, B.D.; Maruniak, J.E. Phylogenetic interrelationships among baculoviruses: Evolutionary rates and host associations. J. Invertebr. Pathol. 1993, 62, 147–164. [Google Scholar] [CrossRef]
- Huang, N.; Civciristov, S.; Hawkins, C.J.; Clem, R.J. Sf-Dronc, an initiator caspase involved in apoptosis in the fall armyworm Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2013, 43, 444–454. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhou, K.; Liu, H.; Wu, A.; Mei, L.; Liu, Q. SfDredd, a Novel Initiator Caspase Possessing Activity on Effector Caspase Substrates in Spodoptera frugiperda. PLoS ONE 2016, 11, e151016. [Google Scholar] [CrossRef] [PubMed]
- Dorstyn, L.; Colussi, P.A.; Quinn, L.M.; Richardson, H.; Kumar, S. DRONC, an ecdysone-inducible drosophila caspase. Proc. Natl. Acad. Sci. USA 1999, 96, 4307–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Song, J.; Bao, X.Y.; Chen, P.; Yi, H.S.; Pan, M.H.; Lu, C. Bmdredd is an initiator caspase and participates in emodin-induced apoptosis in the silkworm, Bombyx mori. Gene 2016, 591, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.M.; Pio, F.; Thi, E.P.; Theilmann, D.; Lowenberger, C. Characterization of Aedes Dredd: A novel initiator caspase from the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2007, 37, 559–569. [Google Scholar] [CrossRef]
- Hu, S.; Yang, X. dFADD, a Novel Death Domain-containing Adapter Protein for the Drosophila Caspase DREDD. J. Biol. Chem. 2000, 275, 30761–30764. [Google Scholar] [CrossRef]
- Yao, L.G.; Liu, Z.C.; Zhang, X.M.; Kan, Y.C.; Zhou, J.J. A highly efficient method for the generation of a recombinant Bombyx mori nuclear-polyhedrosis-virus Bacmid and large-scale expression of foreign proteins in silkworm (B. mori) larvae. Biotechnol. Appl. Biochem. 2007, 48, 45–53. [Google Scholar]
- Sun, J.C.; Zhang, E.H.; Yao, L.G.; Zhang, H.L.; Jin, P.F. A highly efficient method of constructing recombinant Bombyx mori (silkworm) multiple nucleopolyhedrovirus based on zero-background Tn7-mediated transposition in Escherichia coli. Biotechnol. Prog. 2009, 25, 524–529. [Google Scholar] [CrossRef]
- Yao, L.; Sun, J.; Xu, H.; Kan, Y.; Zhang, X.; Yan, H.C. A novel economic method for high throughput pro-duction of recombinant baculovirus by infecting insect cells with Bac-mid-containing diminopimelate-auxotrophic Escherichia coli. J. Biotechnol. 2010, 145, 23–29. [Google Scholar] [CrossRef]
- Yao, L.; Wang, S.; Su, S.; Yao, N.; He, J.; Peng, L.; Sun, J. Construction of a baculovirus-silkworm multigene expression system and its application on producing virus-like particles. PLoS ONE 2012, 7, e32510. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Wang, X.; Ren, F.; Zou, S.; Feng, M.; Xu, L.; Yao, L.; Sun, J. Construction of a highly efficient display system for baculovirus and its application on multigene co-display. Mol. Genet. Genom. 2018, 293, 1265–1277. [Google Scholar] [CrossRef]
- Lock, M.; Korn, M.; Wilson, J.; Sena-Esteves, M.; Gao, G. Measuring the Infectious Titer of Recombinant Adenovirus Using Tissue Culture Infection Dose 50% (TCID50) End-Point Dilution and Quantitative Polymerase Chain Reaction (qPCR). Cold Spring Harb. Protoc. 2019, 2019, pdb.prot095562. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Pan, Y.; Wang, X.; Tian, W.; Yao, L.; Sun, J. Impact of Molecular Modification on the Efficiency of Recombinant Baculovirus Vector Invasion to Mammalian Cells and Its Immunogenicity in Mice. Viruses 2022, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Jiang, Y.; Fan, W.; Yang, C.; Qi, Y. Structural comparisons of capsid protein VP39 and evolution of insect viruses. J. Cent. China Norm. Univ. 2003, 37, 376–381. [Google Scholar]
- Katsuma, S.; Kokusho, R.; Mcfadden, G. A Conserved Glycine Residue Is Required for Proper Functioning of a Baculovirus VP39 Protein. J. Virol. 2017, 91, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birnbaum, M.J.; Clem, R.J.; Miller, L.K. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 1994, 68, 2521–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seshagiri, S.; Miller, L. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc. Natl. Acad. Sci. USA 1997, 94, 13606–13611. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef]
- Jackson, S.; Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 2013, 49, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Vilaplana, L.; O’Reilly, D.R. Functional interaction between Cydia pomonella granulovirus IAP proteins. Virus Res. 2003, 92, 107–111. [Google Scholar] [CrossRef]
- Everett, H.; Mcfadden, G. Viruses and Apoptosis: Meddling with Mitochondria. Virology 2001, 288, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.H.; Deveraux, Q.L.; Maeda, S.; Salvesen, G.S.; Stennicke, H.R.; Hammock, B.D.; Reed, J.C. Evolutionary conservation of apoptosis mechanisms: Lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 2000, 97, 1427–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, M.C.; Monser, K.P.; Clem, R.J. Ubiquitin protein ligase activity of the anti-apoptotic baculovirus protein Op-IAP3. Virus Res. 2004, 105, 89–96. [Google Scholar] [CrossRef]
- Nai, Y.S.; Yang, Y.T.; Kim, J.S.; Wu, C.Y.; Chen, Y.W.; Wang, C.H. Baculoviral iap2 and iap3 encoded by Lymantria xylina multiple nucleopolyhedrovirus (lyxymnpv) suppress insect cell apoptosis in a transient expression assay. Appl. Entomol. Zool. 2016, 51, 305–316. [Google Scholar] [CrossRef]
- Cheung, C.; Chang, Y.C.; Lin, T.Y.; Cheng, S.M.; Leung, E. Anti-apoptotic proteins in the autophagic world: An update on functions of XIAP, Survivin, and BRUCE. J. Biomed. Sci. 2020, 27, 31. [Google Scholar] [CrossRef]
- Silverman, N.S.; Maniatis, T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001, 15, 2321–2342. [Google Scholar] [CrossRef] [Green Version]
- Goshima, T.; Shimada, M.; Sharif, J.; Matsuo, H.; Misaki, T.; Johmura, Y.; Murata, K.; Koseki, H.; Nakanishi, M. Mammal-specific H2A Variant. H2ABbd, Is Involved in Apoptotic Induction via Activation of NF-κB Signaling Pathway. J. Biol. Chem. 2014, 289, 11656–11666. [Google Scholar] [CrossRef] [Green Version]
- Migneault, F.; Dieudé, M.; Turgeon, J. Apoptotic exosome-like vesicles regulate endothelial gene expression, inflammatory signaling, and function through the NF-κB signaling pathway. Sci. Rep. 2020, 10, 12562. [Google Scholar] [CrossRef]
- Nai, Y.S.; Wu, C.Y.; Wang, T.C.; Chen, Y.R.; Lau, W.H.; Lo, C.F.; Tsai, M.F.; Wang, C.H. Genomic sequencing and analyses of Lymantria xylina multiple nucleopolyhedrovirus. BMC Genom. 2010, 11, 116. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.C.; Doctor, K.S.; Godzik, A. The Domains of Apoptosis: A Genomics Perspective. Sci. Signal. 2004, 2004, re9. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.W.; Clem, R.J. Sequence requirements for Hid binding and apoptosis regulation in the baculovirus inhibitor of apoptosis Op-IAP. Hid binds Op-IAP in a manner similar to Smac binding of XIAP. J. Biol. Chem. 2002, 277, 2454–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenev, T.; Zachariou, A.; Wilson, R.; Ditzel, M.; Meier, P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat. Cell Biol. 2005, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Servi, B.D.; Hermani, A.; Medunjanin, S.; Mayer, D. Impact of PKCδ on estrogen receptor localization and activity in breast cancer cells. Oncogene 2005, 24, 4946–4955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soroush, F.; Tang, Y.; Zaidi, H.M.; Sheffield, J.B. Kilpatrick LE. PKCδ inhibition as a novel medical countermeasure for radiation-induced vascular damage. FASEB J. 2018, 32, 6436–6444. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Becnel, J.J.; Zhang, Y. Induction of reaper ortholog mx in mosquito midgut cells following baculovirus infection. Cell Death Differ. 2011, 18, 1337. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wu, Y.; Hui, T.; Hui, L. Reaper homologue IBM1 in silkworm Bombyx mori induces apoptosis upon baculovirus infection. FEBS Lett. 2013, 587, 600–606. [Google Scholar] [CrossRef] [Green Version]
- Deveraux, Q.L.; Reed, J.C. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999, 13, 239–252. [Google Scholar] [CrossRef]
| Name | Sequence | Size (bp) |
|---|---|---|
| Bsiap1 (with His-Tag) | F: 5′-aggatccatgcatcatcaccaccatcacatgtgttgttttttttggc R: 5′-agagctcttaatttacatacaatttttg | 549 |
| Bsiap2 (with V5-Tag) | F: 5′-aggatccatgggtaagcctatccctaaccctctcctcggtctcgattctacgatgaactacgaaagtgct R: 5′-agagctcttataaaaacactttaatc | 933 |
| Bsiap3 (with Flag-Tag) | F: 5′-aggatccatggattacaaggatgacgacgataagatgtacatagaagatt R: 5′-agagctcttacacttgatacatgc | 831 |
| gfp | F: 5′-acccgggatggtgagcaagggcgagga R: 5′-agctagcttacttgtacagctcgtccat | 720 |
| mCherry | F: 5′-acccgggatggtgagcaagggcgagga R: 5′-agctagcttacttgtacagctcgtccatgcc | 711 |
| Gene | Primer Sequence | Gene ID | Size (bp) |
|---|---|---|---|
| α-Tubulin | F: 5′-agtccagatcggtaatgc R: 5′-gctgaagaaggtgttgaag | ABX55885.1 | 124 |
| baculovirus | F: 5′-cccgtaacggacctcgtactt R: 5′-ttatcgagatttatttgcatacaac | NC_001623.1 | 141 |
| caspase-1 | F: 5′-gattcaaagttacggtgttcccta R: 5′-ggttgtctggcttgtaatgagtat | U81510.1 | 171 |
| caspase-2 | F: 5′-gtaaggttctgattggcaattagc R: 5′-cggtacttgtggttggtgtt | KP711808.1 | 173 |
| caspase-9 | F: 5′-acacagagtttgacaacaatatcg R: 5′-ggtctcatagtccaccaacac | KC683711.1 | 179 |
| dronc | F: 5′-ctggtagatacgcttggagaacta R: 5′-gcctgtttgatgtgctaagact | JX912275.1 | 160 |
| dredd | F: 5′-aacaccacaaggaatggaagt R: 5′-agttacaggcatcgttggaa | KU668855.1 | 200 |
| Sf9-iap | F: 5′-gttggagagttgtgttgtttgttt R: 5′-aatagcgttaatgttgaggaggag | AF186378.1 | 199 |
| vp39 | F: 5′-acccgataagaagcagtgac R: 5′-cccagagtagcgttaggatt | NC_001623.1 | 226 |
| p35 | F: 5′-cgaacgcaacgactactac R: 5′-tgagcaaacggcacaataac | NC_001623.1 | 125 |
| p53 | F: 5′-caccgtctcaaccgtatc R: 5′-gaggacattcttcgctattt | HM773026.1 | 210 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, H.; Pan, Y.; Awais, M.M.; Tian, W.; Li, J.; Sun, J. Impact of Group II Baculovirus IAPs on Virus-Induced Apoptosis in Insect Cells. Genes 2022, 13, 750. https://doi.org/10.3390/genes13050750
Zheng H, Pan Y, Awais MM, Tian W, Li J, Sun J. Impact of Group II Baculovirus IAPs on Virus-Induced Apoptosis in Insect Cells. Genes. 2022; 13(5):750. https://doi.org/10.3390/genes13050750
Chicago/Turabian StyleZheng, Hao, Yong Pan, Mian Muhammad Awais, Weibin Tian, Jingyang Li, and Jingchen Sun. 2022. "Impact of Group II Baculovirus IAPs on Virus-Induced Apoptosis in Insect Cells" Genes 13, no. 5: 750. https://doi.org/10.3390/genes13050750






