Transcriptomic Analysis of Canine Osteosarcoma from a Precision Medicine Perspective Reveals Limitations of Differential Gene Expression Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of Data
2.2. RNA Isolation and Sequencing
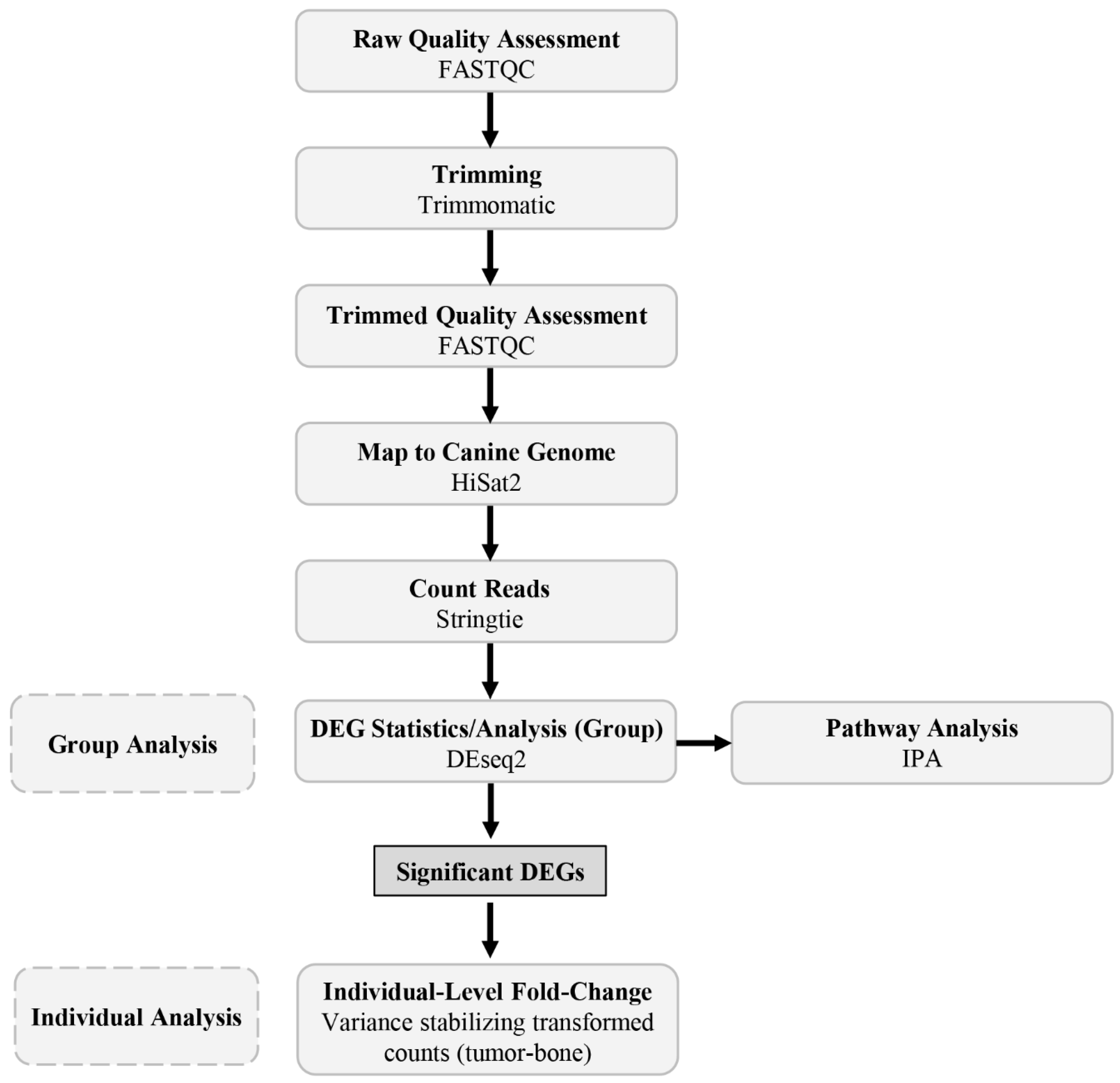
2.3. Bioinformatic Pipeline and Data Analysis
2.4. Group Analysis
2.5. Individual Analysis
3. Results
3.1. RNA Quality
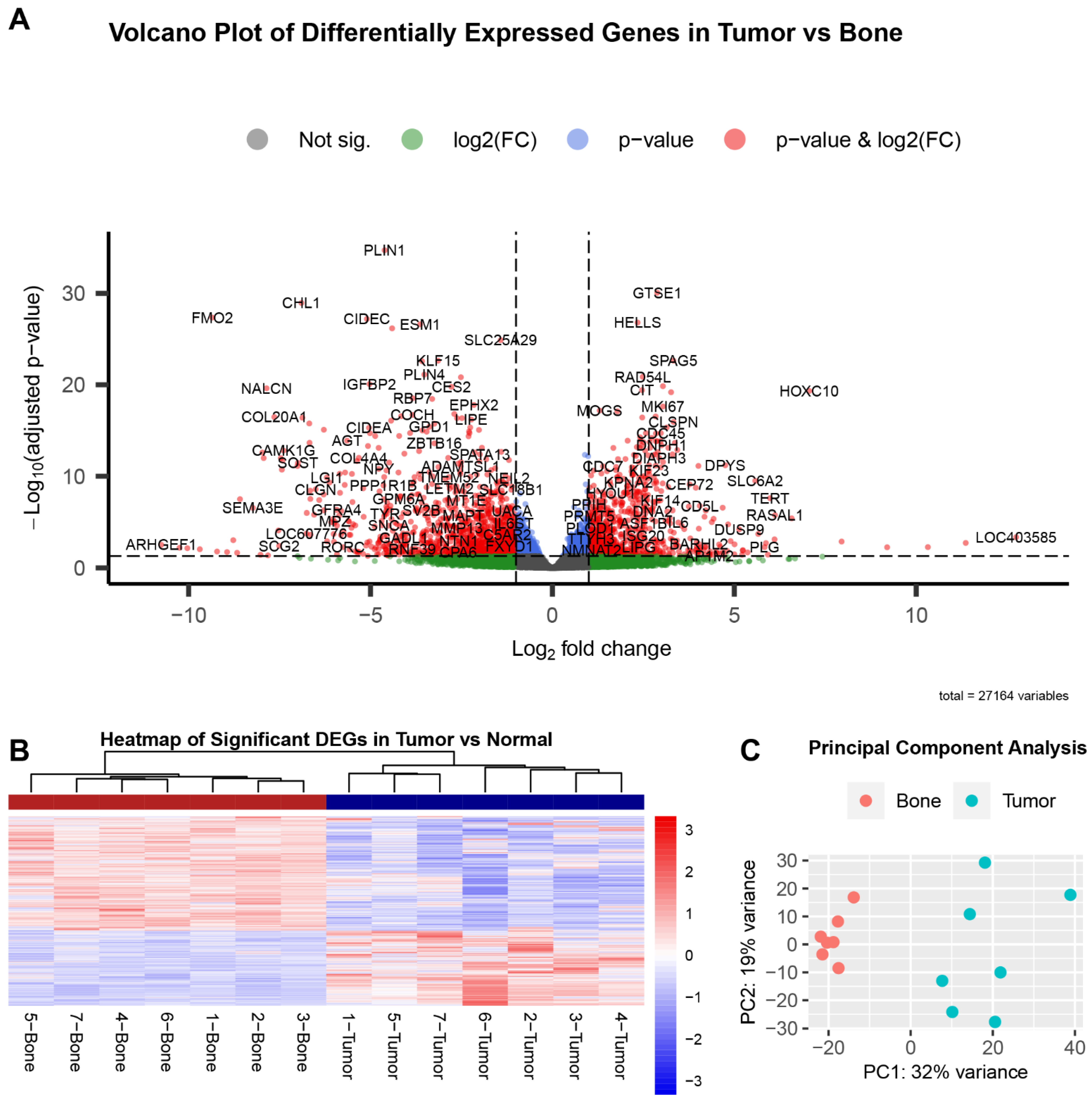
3.2. Group Analysis
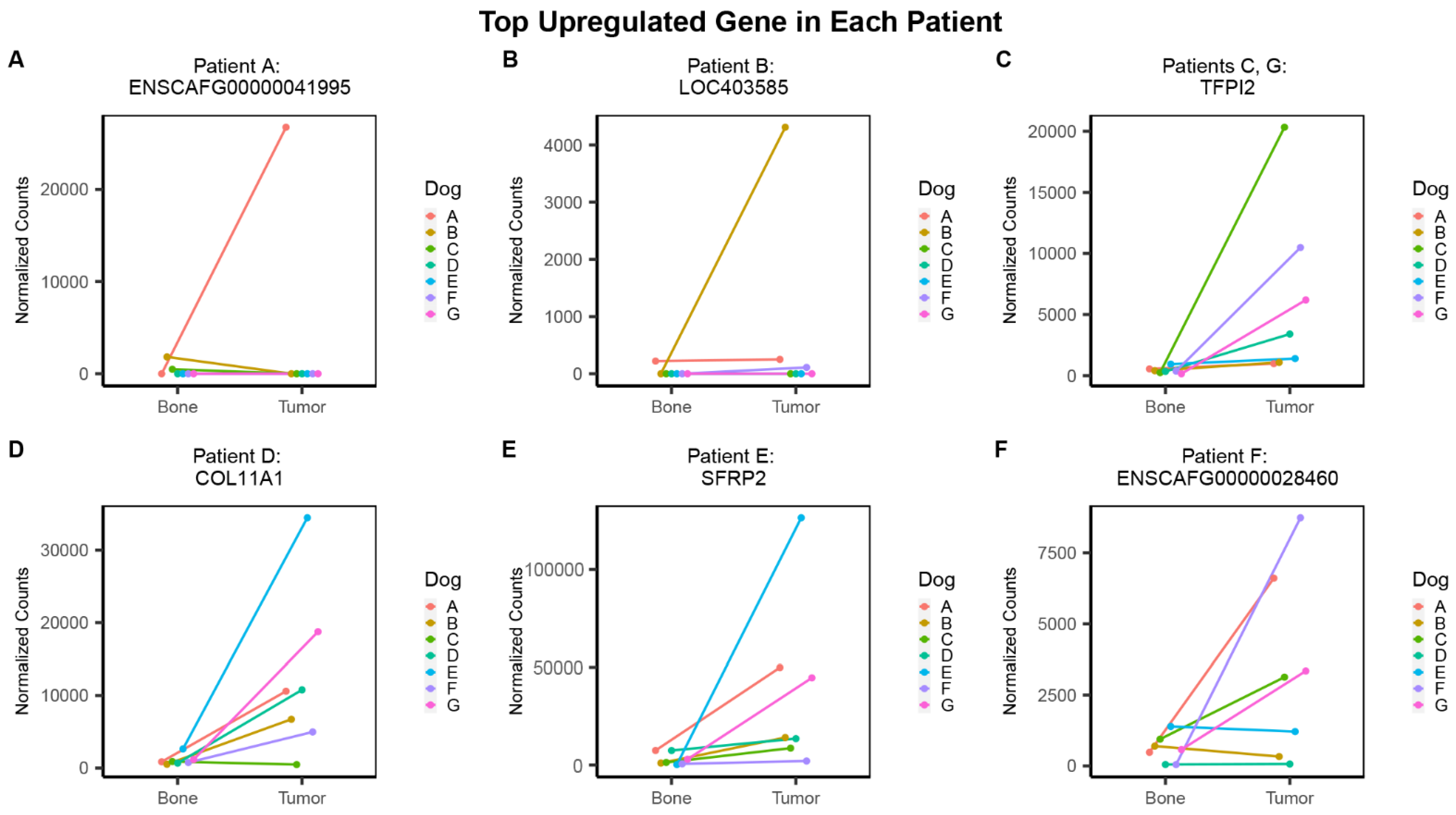
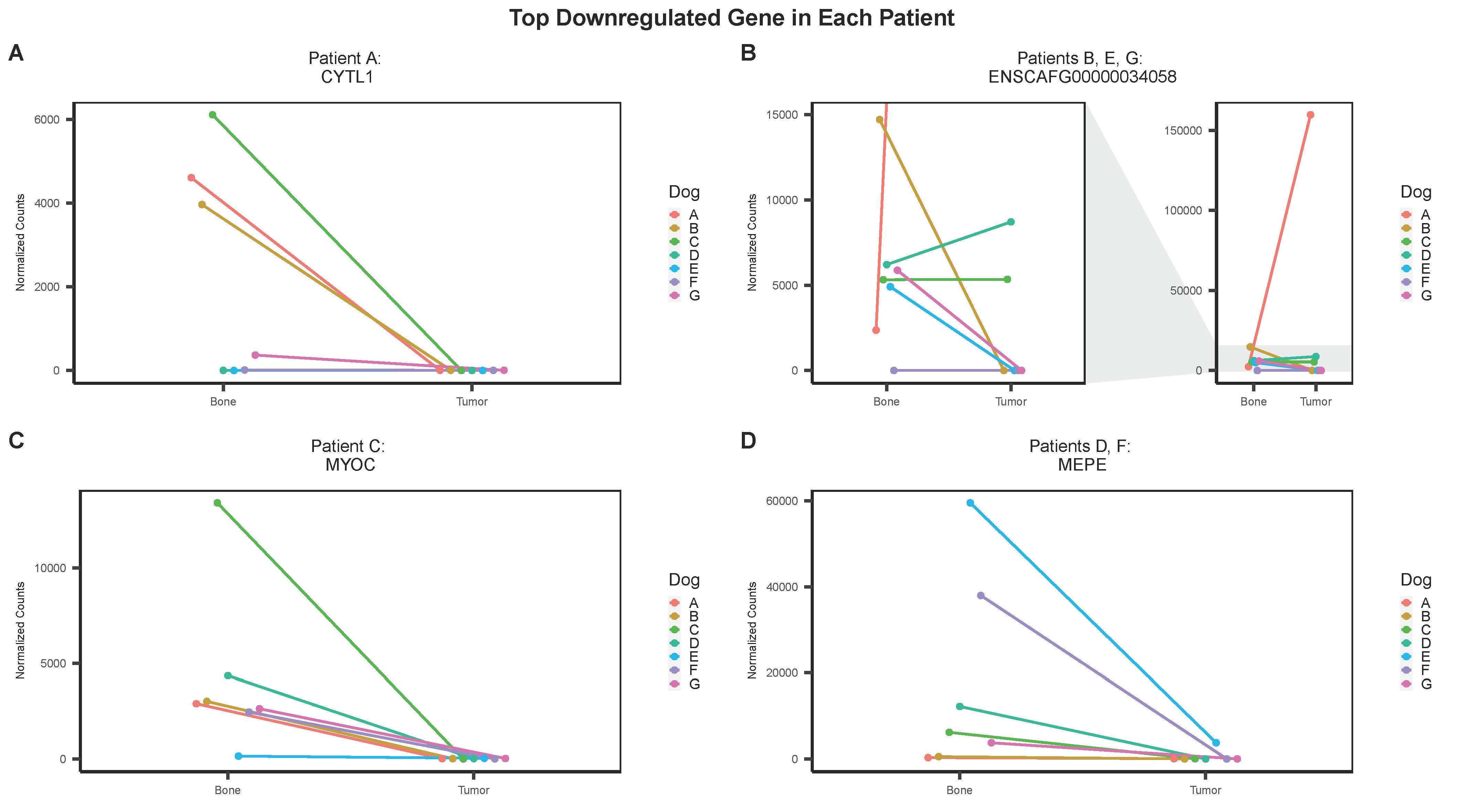
3.3. Individual Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geller, D.S.; Gorlick, R. Osteosarcoma: A Review of Diagnosis, Management, and Treatment Strategies. Clin. Adv. Hematol. Oncol. 2010, 8, 705–718. [Google Scholar] [PubMed]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma Treatment—Where Do We Stand? A State of the Art Review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Longhi, A.; Errani, C.; De Paolis, M.; Mercuri, M.; Bacci, G. Primary Bone Osteosarcoma in the Pediatric Age: State of the Art. Cancer Treat. Rev. 2006, 32, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Gorlick, R.; Khanna, C. Osteosarcoma. J. Bone Min. Res. 2010, 25, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma Incidence and Survival Rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef]
- Sayles, L.C.; Breese, M.R.; Koehne, A.L.; Leung, S.G.; Lee, A.G.; Liu, H.-Y.; Spillinger, A.; Shah, A.T.; Tanasa, B.; Straessler, K.; et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019, 9, 46–63. [Google Scholar] [CrossRef]
- Paoloni, M.; Davis, S.; Lana, S.; Withrow, S.; Sangiorgi, L.; Picci, P.; Hewitt, S.; Triche, T.; Meltzer, P.; Khanna, C. Canine Tumor Cross-Species Genomics Uncovers Targets Linked to Osteosarcoma Progression. BMC Genom. 2009, 10, 625. [Google Scholar] [CrossRef]
- Sakthikumar, S.; Elvers, I.; Kim, J.; Arendt, M.L.; Thomas, R.; Turner-Maier, J.; Swofford, R.; Johnson, J.; Schumacher, S.E.; Alföldi, J.; et al. SETD2 Is Recurrently Mutated in Whole-Exome Sequenced Canine Osteosarcoma. Cancer Res. 2018, 78, 3421–3431. [Google Scholar] [CrossRef]
- Gardner, H.L.; Sivaprakasam, K.; Briones, N.; Zismann, V.; Perdigones, N.; Drenner, K.; Facista, S.; Richholt, R.; Liang, W.; Aldrich, J.; et al. Canine Osteosarcoma Genome Sequencing Identifies Recurrent Mutations in DMD and the Histone Methyltransferase Gene SETD2. Commun. Biol. 2019, 2, 266. [Google Scholar] [CrossRef]
- Withrow, S.J. Withrow and MacEwen’s Small Animal Clinical Oncology; Elsevier Health Sciences: St. Louis, MO, USA, 2007; ISBN 978-0-7216-0558-6. [Google Scholar]
- Simpson, S.; Dunning, M.D.; de Brot, S.; Grau-Roma, L.; Mongan, N.P.; Rutland, C.S. Comparative Review of Human and Canine Osteosarcoma: Morphology, Epidemiology, Prognosis, Treatment and Genetics. Acta Vet. Scand. 2017, 59, 71. [Google Scholar] [CrossRef]
- Zhao, J.; Dean, D.C.; Hornicek, F.J.; Yu, X.; Duan, Z. Emerging Next-Generation Sequencing-Based Discoveries for Targeted Osteosarcoma Therapy. Cancer Lett. 2020, 474, 158–167. [Google Scholar] [CrossRef]
- Martin, J.W.; Squire, J.A.; Zielenska, M. The Genetics of Osteosarcoma. Sarcoma 2012, 2012, e627254. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, K.A.; Barber, L.J.; Davies, M.N.; Ashenden, M.; Sottoriva, A.; Gerlinger, M. Cancer Evolution and the Limits of Predictability in Precision Cancer Medicine. Trends Cancer 2016, 2, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Wagner, M.J.; McGuire, M.F.; Sarwari, N.M.; Devarajan, E.; Lewis, V.O.; Westin, S.; Kato, S.; Brown, R.E.; Anderson, P. Personalized Comprehensive Molecular Profiling of High Risk Osteosarcoma: Implications and Limitations for Precision Medicine. Oncotarget 2015, 6, 40642–40654. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.; Dunning, M.; de Brot, S.; Alibhai, A.; Bailey, C.; Woodcock, C.L.; Mestas, M.; Akhtar, S.; Jeyapalan, J.N.; Lothion-Roy, J.; et al. Molecular Characterisation of Canine Osteosarcoma in High Risk Breeds. Cancers 2020, 12, 2405. [Google Scholar] [CrossRef]
- Endo-Munoz, L.; Cumming, A.; Sommerville, S.; Dickinson, I.; Saunders, N.A. Osteosarcoma Is Characterised by Reduced Expression of Markers of Osteoclastogenesis and Antigen Presentation Compared with Normal Bone. Br. J. Cancer 2010, 103, 73–81. [Google Scholar] [CrossRef]
- Xie, L.; Yao, Z.; Zhang, Y.; Li, D.; Hu, F.; Liao, Y.; Zhou, L.; Zhou, Y.; Huang, Z.; He, Z.; et al. Deep RNA Sequencing Reveals the Dynamic Regulation of MiRNA, LncRNAs, and MRNAs in Osteosarcoma Tumorigenesis and Pulmonary Metastasis. Cell Death Dis. 2018, 9, 772. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Q.; Lin, F.; Wang, X.; Wang, Y.; Wang, J.; Wang, C. RNA Sequencing of Osteosarcoma Gene Expression Profile Revealed That MiR-214-3p Facilitates Osteosarcoma Cell Proliferation via Targeting Ubiquinol-Cytochrome c Reductase Core Protein 1 (UQCRC1). Med. Sci. Monit. 2019, 25, 4982–4991. [Google Scholar] [CrossRef]
- Glickman, M.S.; Sawyers, C.L. Converting Cancer Therapies into Cures: Lessons from Infectious Diseases. Cell 2012, 148, 1089–1098. [Google Scholar] [CrossRef]
- Martson, A.; Kõks, S.; Reimann, E.; Prans, E.; Erm, T.; Maasalu, K. Transcriptome Analysis of Osteosarcoma Identifies Suppression of Wnt Pathway and Up-Regulation of Adiponectin as Potential Biomarker. Genom. Discov. 2013, 1, 3. [Google Scholar] [CrossRef][Green Version]
- Ho, X.D.; Phung, P.; Q Le, V.; H Nguyen, V.; Reimann, E.; Prans, E.; Kõks, G.; Maasalu, K.; Le, N.T.; H Trinh, L.; et al. Whole Transcriptome Analysis Identifies Differentially Regulated Networks between Osteosarcoma and Normal Bone Samples. Exp. Biol. Med. 2017, 242, 1802–1811. [Google Scholar] [CrossRef]
- Nance, R.; Agarwal, P.; Sandey, M.; Starenki, D.; Koehler, J.; Sajib, A.M.; Smith, B.F. A Method for Isolating RNA from Canine Bone. Biotechniques 2020, 68, 311–317. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 October 2021).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Rapp, N.; Deans, R.; Cheng, L. Molecular Cloning and Chromosomal Mapping of a Candidate Cytokine Gene Selectively Expressed in Human CD34+ Cells. Genomics 2000, 65, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Dugger, S.A.; Platt, A.; Goldstein, D.B. Drug Development in the Era of Precision Medicine. Nat. Rev. Drug Discov. 2018, 17, 183–196. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor Heterogeneity: Causes and Consequences. Biochim. Biophys. Acta 2010, 1805, 105. [Google Scholar] [CrossRef]
- Lin, F.; Xie, Y.-J.; Zhang, X.-K.; Huang, T.-J.; Xu, H.-F.; Mei, Y.; Liang, H.; Hu, H.; Lin, S.-T.; Luo, F.-F.; et al. GTSE1 Is Involved in Breast Cancer Progression in P53 Mutation-Dependent Manner. J. Exp. Clin. Cancer Res. 2019, 38, 152. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhu, W.; Li, X.; Han, Y.; Wang, Y.; Leng, Q.; Li, M.; Wen, X. GTSE1 Promotes Prostate Cancer Cell Proliferation via the SP1/FOXM1 Signaling Pathway. Lab. Investig. 2021, 101, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, H.; Lian, Y.; Chen, L.; Gu, L.; Wang, J.; Huang, Y.; Deng, M.; Gao, Z.; Huang, Y. GTSE1 Promotes Cell Migration and Invasion by Regulating EMT in Hepatocellular Carcinoma and Is Associated with Poor Prognosis. Sci. Rep. 2017, 7, 5129. [Google Scholar] [CrossRef]
- Liu, A.; Zeng, S.; Lu, X.; Xiong, Q.; Xue, Y.; Tong, L.; Xu, W.; Sun, Y.; Zhang, Z.; Xu, C. Overexpression of G2 and S Phase-Expressed-1 Contributes to Cell Proliferation, Migration, and Invasion via Regulating P53/FoxM1/CCNB1 Pathway and Predicts Poor Prognosis in Bladder Cancer. Int. J. Biol. Macromol. 2019, 123, 322–334. [Google Scholar] [CrossRef]
- Xie, C.; Xiang, W.; Shen, H.; Shen, J. GTSE1 Is Possibly Involved in the DNA Damage Repair and Cisplatin Resistance in Osteosarcoma. J. Orthop. Surg. Res. 2021, 16, 713. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, M.; Zhou, L.; Zhang, Y.; Liu, W.; Qin, W.; He, R.; Lu, Y.; Wang, Y.; Chen, X.-Z.; et al. Prognostic Significance of PLIN1 Expression in Human Breast Cancer. Oncotarget 2016, 7, 54488–54502. [Google Scholar] [CrossRef]
- Lu, C.; Wang, X.; Zhao, X.; Xin, Y.; Liu, C. Long Non-Coding RNA ARAP1-AS1 Accelerates Cell Proliferation and Migration in Breast Cancer through MiR-2110/HDAC2/PLIN1 Axis. Biosci. Rep. 2020, 40, BSR20191764. [Google Scholar] [CrossRef]
- Jung, H.; Yoon, S.R.; Lim, J.; Cho, H.J.; Lee, H.G. Dysregulation of Rho GTPases in Human Cancers. Cancers 2020, 12, 1179. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhong, Y.; Li, X.; Du, S.; Xie, P.; Zheng, G.; Han, J. Identification of Co-Expression Modules and Pathways Correlated with Osteosarcoma and Its Metastasis. World J. Surg. Oncol. 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yoo, S.; Zhou, R.; Xu, A.; Bernitz, J.M.; Yuan, Y.; Gomes, A.M.; Daniel, M.G.; Su, J.; Demicco, E.G.; et al. Oncogenic Role of SFRP2 in P53-Mutant Osteosarcoma Development via Autocrine and Paracrine Mechanism. Proc. Natl. Acad. Sci. USA 2018, 115, E11128–E11137. [Google Scholar] [CrossRef] [PubMed]
- Sierko, E.; Wojtukiewicz, M.Z.; Kisiel, W. The Role of Tissue Factor Pathway Inhibitor-2 in Cancer Biology. Semin. Thromb. Hemost. 2007, 33, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, C.; Wang, S.; Xiang, Z.; Dou, R.; Lin, Z.; Zheng, J.; Xiong, B. SDC2 and TFPI2 Methylation in Stool Samples as an Integrated Biomarker for Early Detection of Colorectal Cancer. Cancer Manag. Res. 2021, 13, 3601–3617. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xiao, X.; Zhou, X.; Huang, T.; Du, C.; Yu, N.; Mo, Y.; Lin, L.; Zhang, J.; Ma, N.; et al. TFPI-2 Is a Putative Tumor Suppressor Gene Frequently Inactivated by Promoter Hypermethylation in Nasopharyngeal Carcinoma. BMC Cancer 2010, 10, 617. [Google Scholar] [CrossRef] [PubMed]


| Patient | Sex | Neuter Status | Age | Tumor Site | Breed | Tumor RIN | Bone RIN |
|---|---|---|---|---|---|---|---|
| A | M | Castrated | 10 yrs, 6 mos | Left distal radius | Golden Retriever | 5.0 | 6.6 |
| B | M | Castrated | 7 yrs | Left distal femur | Rottweiler | 7.4 | 4.5 |
| C | M | Castrated | 7 yrs | Left distal radius | Doberman Pinscher | 6.9 | 6.5 |
| D | M | Castrated | 11 yrs, 9 mos | Left proximal humerus | Greyhound | 6.4 | 6.7 |
| E | F | Spayed | 7 yrs, 6 mos | Left distal radius | Great Pyrenees | 6.9 | 6.9 |
| F | M | Castrated | 7 yrs | Right distal radius | Golden Retriever | 8.3 | 7.1 |
| G | F | Spayed | 9 yrs, 9 mos | Right distal tibia | Greyhound | 6.7 | 7.2 |
| Top 10 Downregulated Genes in Tumor, Ordered by Log2 Fold-Change | ||||
|---|---|---|---|---|
| ENSEMBL | SYMBOL | Log2FC | Padj | GENE NAME |
| ENSCAFG00000028799 | ARHGEF1 | −10.75 | 2.93 × 10−3 | Rho guanine nucleotide exchange factor 1 |
| ENSCAFG00000041995 | NA | −10.25 | 6.33 × 10−3 | NA |
| ENSCAFG00000007622 | NA | −10.04 | 6.95 × 10−3 | NA |
| ENSCAFG00000005350 | NA | −9.69 | 9.07 × 10−3 | NA |
| ENSCAFG00000014986 | FMO2 | −9.35 | 4.40 × 10−28 | flavin containing dimethylaniline monoxygenase 2 |
| ENSCAFG00000042006 | NA | −9.22 | 1.63 × 10−2 | NA |
| ENSCAFG00000049609 | NA | −8.94 | 1.84 × 10−2 | NA |
| ENSCAFG00000008109 | NA | −8.77 | 9.28 × 10−4 | NA |
| ENSCAFG00000029213 | LOC607979 | −8.67 | 2.22 × 10−2 | eukaryotic translation initiation factor 3, subunit L pseudogene |
| ENSCAFG00000011465 | NA | −8.59 | 3.20 × 10−8 | NA |
| Top 10 Upregulated Genes in Tumor, Ordered by Log2 Fold-Change | ||||
| ENSCAFG00000044295 | NA | 13.49 | 1.31 × 10−4 | NA |
| ENSCAFG00000009135 | LOC403585 | 12.75 | 4.34 × 10−4 | serum amyloid A1 |
| ENSCAFG00000032019 | NLRP12 | 11.36 | 1.85 × 10−3 | NLR family pyrin domain containing 12 |
| ENSCAFG00000013213 | NA | 10.32 | 5.31 × 10−3 | NA |
| ENSCAFG00000043115 | NA | 9.20 | 5.53 × 10−3 | NA |
| ENSCAFG00000007173 | NA | 7.95 | 1.31 × 10−3 | NA |
| ENSCAFG00000006648 | HOXC10 | 7.06 | 4.67 × 10−3 | homeobox C10 |
| ENSCAFG00000015211 | APOBEC3Z1 | 6.58 | 3.81 × 10−6 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like |
| ENSCAFG00000008986 | RASAL1 | 6.12 | 1.68 × 10−6 | RAS protein activator like 1 |
| ENSCAFG00000035513 | LOC111093651 | 6.11 | 7.53 × 10−4 | uncharacterized LOC111093651 |
| Top 10 Downregulated Genes in Tumor, Ordered by Adjusted p-value | ||||
| ENSCAFG00000011986 | PLIN1 | −4.61 | 1.91 × 10−35 | perilipin 1 |
| ENSCAFG00000006248 | CHL1 | −6.90 | 1.11 × 10−29 | cell adhesion molecule L1 like |
| ENSCAFG00000014986 | FMO2 | −9.35 | 4.40 × 10−28 | flavin containing dimethylaniline monoxygenase 2 |
| ENSCAFG00000005266 | CIDEC | −5.10 | 6.25 × 10−28 | cell death inducing DFFA like effector c |
| ENSCAFG00000018381 | ESM1 | −3.63 | 2.38 × 10−27 | endothelial cell specific molecule 1 |
| ENSCAFG00000006392 | ACKR4 | −4.41 | 6.62 × 10−27 | atypical chemokine receptor 4 |
| ENSCAFG00000030764 | SLC25A29 | −1.42 | 1.31 × 10−25 | solute carrier family 25 member 29 |
| ENSCAFG00000003807 | KLF15 | −3.14 | 2.28 × 10−23 | Kruppel-like factor 15 |
| ENSCAFG00000001854 | AQP7 | −3.58 | 2.66 × 10−23 | aquaporin 7 |
| ENSCAFG00000015323 | PLIN4 | −3.52 | 7.60 × 10−22 | perilipin 4 |
| Top 10 Upregulated Genes in Tumor, Ordered by Adjusted p-value | ||||
| ENSCAFG00000000782 | GTSE1 | 2.88 | 8.51 × 10−31 | G2 and S-phase expressed 1 |
| ENSCAFG00000008090 | HELLS | 2.34 | 1.54 × 10−27 | helicase, lymphoid specific |
| ENSCAFG00000018724 | SPAG5 | 3.33 | 2.28 × 10−23 | sperm associated antigen 5 |
| ENSCAFG00000004272 | RAD54L | 2.48 | 1.35 × 10−21 | RAD54 like |
| ENSCAFG00000016616 | IQGAP3 | 3.04 | 1.41 × 10−20 | IQ motif containing GTPase activating protein 3 |
| ENSCAFG00000010114 | CIT | 2.46 | 3.84 × 10−20 | citron rho-interacting serine/threonine kinase |
| ENSCAFG00000006648 | HOXC10 | 7.06 | 4.67 × 10−20 | homeobox C10 |
| ENSCAFG00000016090 | TOP2A | 3.26 | 6.47 × 10−20 | DNA topoisomerase II α |
| ENSCAFG00000013255 | MKI67 | 3.04 | 2.35 × 10−18 | marker of proliferation Ki-67 |
| ENSCAFG00000008478 | MOGS | 1.30 | 6.47 × 10−18 | mannosyl-oligosaccharide glucosidase |
| Patient | Gene | Log2FC (Individual) | Log2FC (Group) | FDR | |||
|---|---|---|---|---|---|---|---|
| Individual Analysis Results | Top Upregulated Gene | A | ENSCAFG00000041995 | 9.03 | Group Analysis Results | −10.25 | 6.39 × 10−3 |
| B | LOC403585 | 6.42 | 12.75 | 4.37 × 10−4 | |||
| C | TFPI2 | 5.92 | 3.23 | 3.60 × 10−6 | |||
| D | COL11A1 | 3.81 | 3.18 | 1.49 × 10−9 | |||
| E | SFRP2 | 8.63 | 3.19 | 7.36 × 10−6 | |||
| F | ENSCAFG00000028460 | 6.09 | 1.79 | 4.15 × 10−2 | |||
| G | TFPI2 | 4.68 | 3.23 | 3.60 × 10−6 | |||
| Top Downregulated Gene | A | CYTL1 | −6.13 | −6.65 | 2.34 × 10−4 | ||
| B | ENSCAFG00000034058 | −8.17 | −5.92 | 3.31 × 10−2 | |||
| C | MYOC | −8.04 | −7.97 | 2.58 × 10−13 | |||
| D | MEPE | −7.33 | −7.47 | 1.06 × 10−12 | |||
| E | ENSCAFG00000034058 | −6.61 | −5.92 | 3.31 × 10−2 | |||
| F | MEPE | −9.05 | −7.47 | 1.06 × 10−12 | |||
| G | ENSCAFG00000034058 | −6.86 | −5.92 | 3.31 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nance, R.L.; Cooper, S.J.; Starenki, D.; Wang, X.; Matz, B.; Lindley, S.; Smith, A.N.; Smith, A.A.; Bergman, N.; Sandey, M.; et al. Transcriptomic Analysis of Canine Osteosarcoma from a Precision Medicine Perspective Reveals Limitations of Differential Gene Expression Studies. Genes 2022, 13, 680. https://doi.org/10.3390/genes13040680
Nance RL, Cooper SJ, Starenki D, Wang X, Matz B, Lindley S, Smith AN, Smith AA, Bergman N, Sandey M, et al. Transcriptomic Analysis of Canine Osteosarcoma from a Precision Medicine Perspective Reveals Limitations of Differential Gene Expression Studies. Genes. 2022; 13(4):680. https://doi.org/10.3390/genes13040680
Chicago/Turabian StyleNance, Rebecca L., Sara J. Cooper, Dmytro Starenki, Xu Wang, Brad Matz, Stephanie Lindley, Annette N. Smith, Ashley A. Smith, Noelle Bergman, Maninder Sandey, and et al. 2022. "Transcriptomic Analysis of Canine Osteosarcoma from a Precision Medicine Perspective Reveals Limitations of Differential Gene Expression Studies" Genes 13, no. 4: 680. https://doi.org/10.3390/genes13040680
APA StyleNance, R. L., Cooper, S. J., Starenki, D., Wang, X., Matz, B., Lindley, S., Smith, A. N., Smith, A. A., Bergman, N., Sandey, M., Koehler, J., Agarwal, P., & Smith, B. F. (2022). Transcriptomic Analysis of Canine Osteosarcoma from a Precision Medicine Perspective Reveals Limitations of Differential Gene Expression Studies. Genes, 13(4), 680. https://doi.org/10.3390/genes13040680








