Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Measurement of Physiological Indices
2.3. Proteomic Analysis of Two Maize Varieties
2.4. RNA Extraction and Quantitative Real-Time PCR(qRT-PCR)
2.5. Bioinformatics Analysis of ZmTGA
2.6. Function Verification of ZmTGA Gene
3. Results
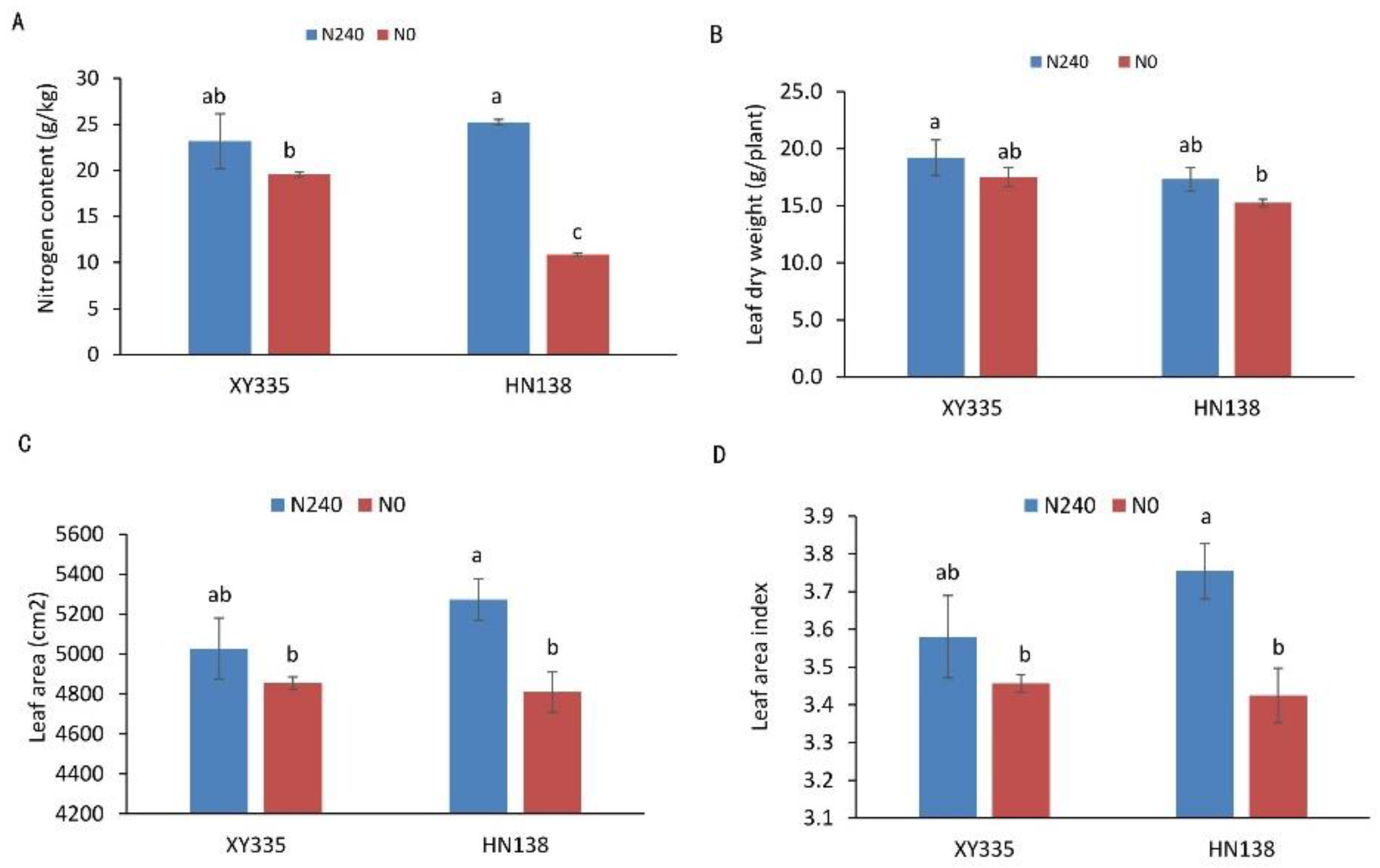
3.1. Phenotypic and Physiological Characteristics of Two Hybrids under Low-Nitrogen Stress
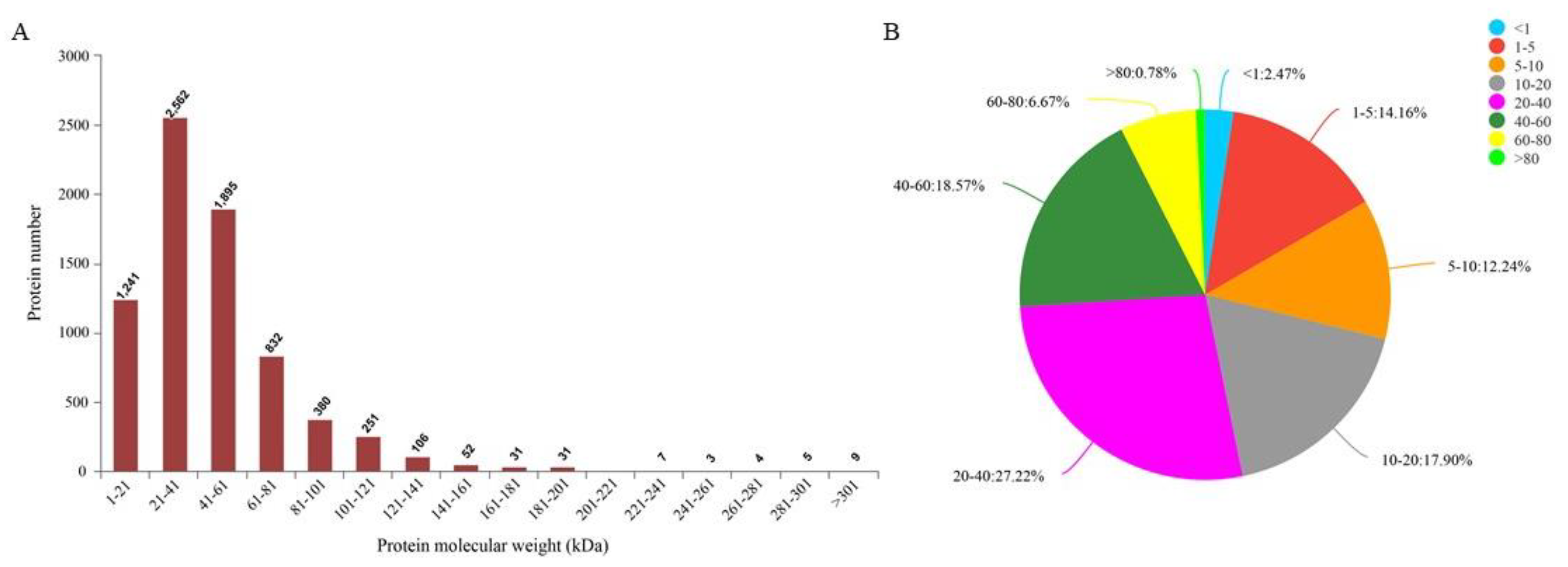
3.2. Overall View of Maize Leaf Proteins Identified by TMT Analysis
3.3. Analysis of DAPs Observed in Different Experimental Comparisons
3.4. Gene Ontology Annotation and Functional Classification
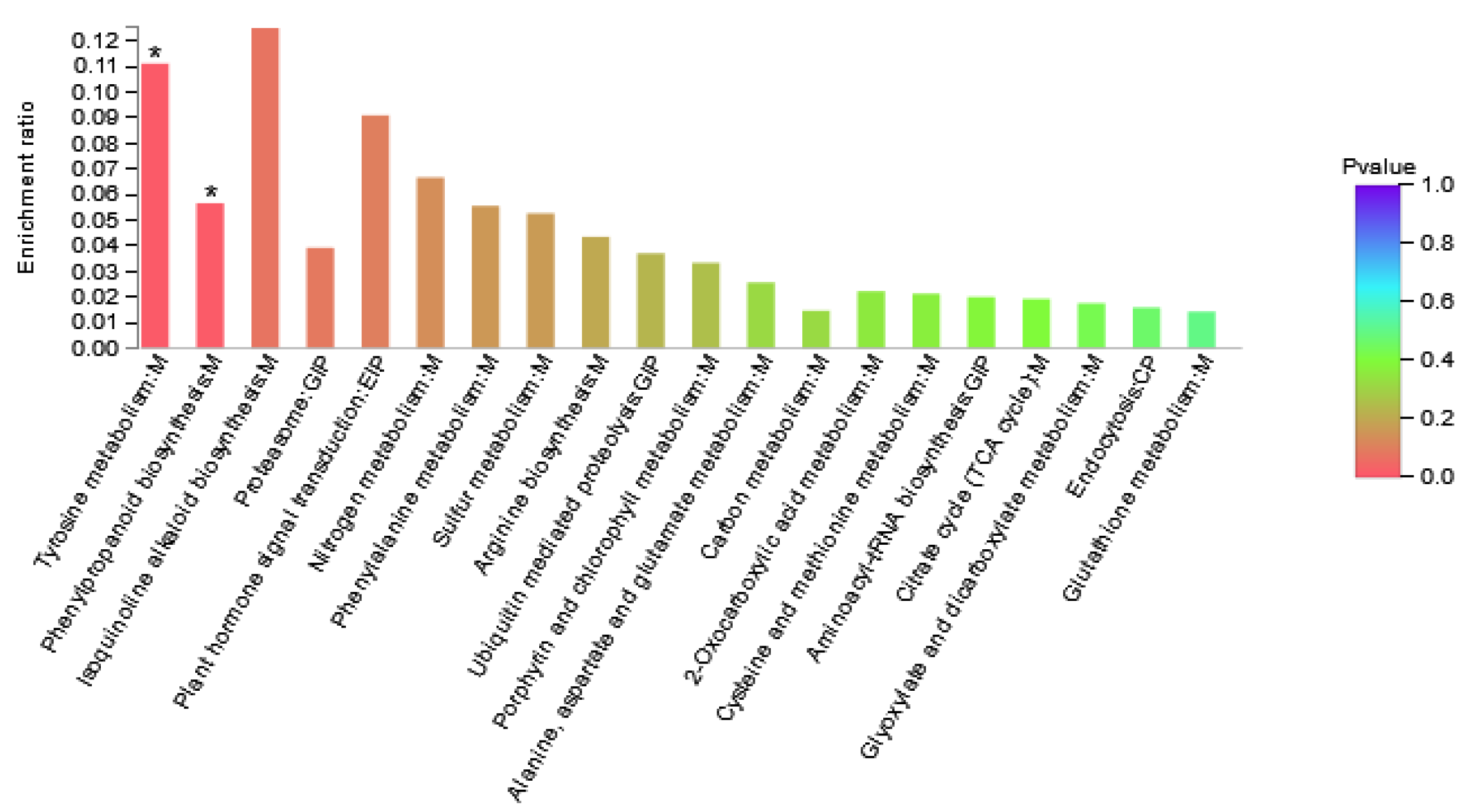
3.5. KEGG Pathway Enrichment Analysis of DAPs
3.6. qRT-PCR Analysis
3.7. Physicochemical Properties and Structure Analysis of ZmTGA
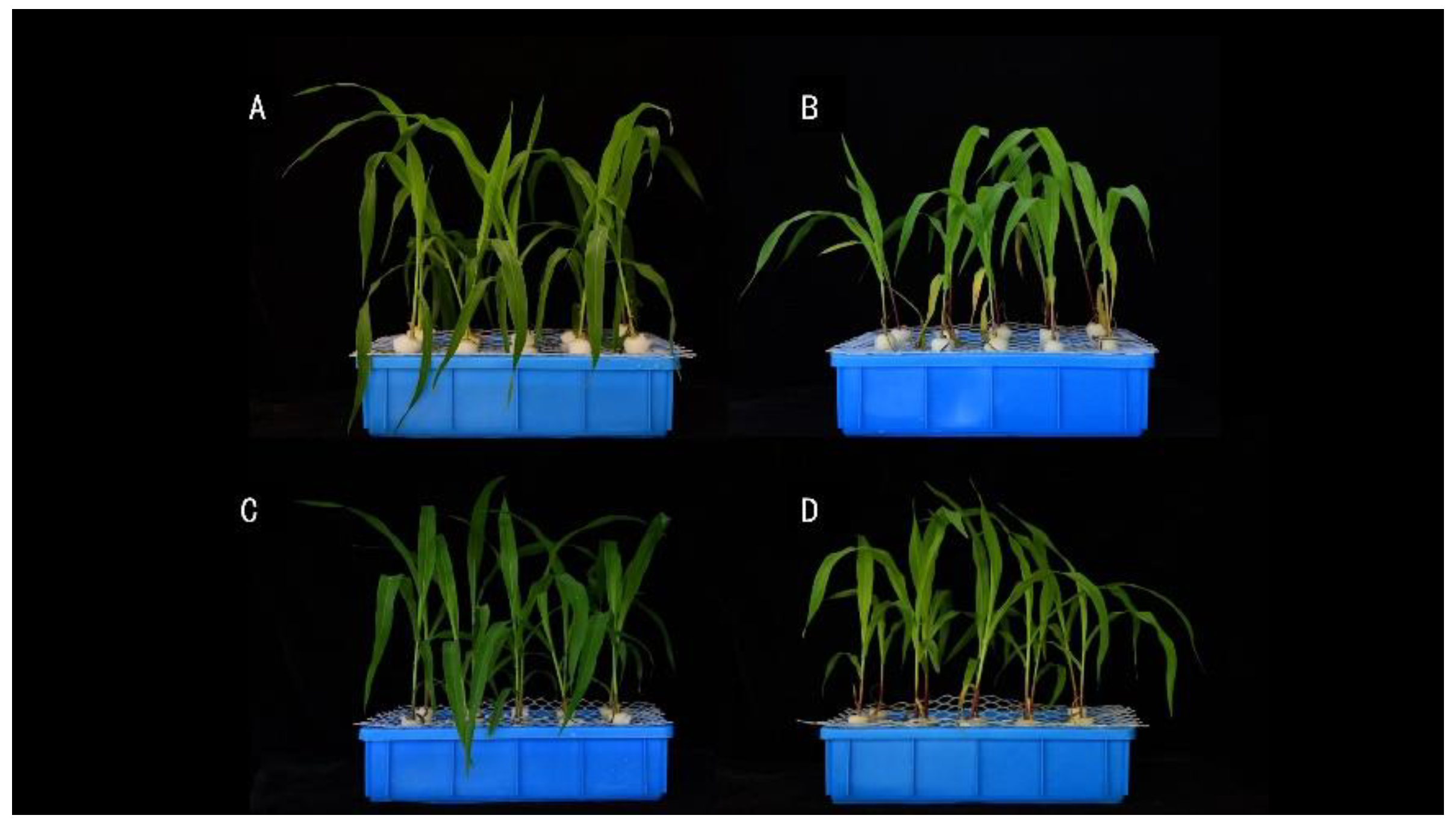
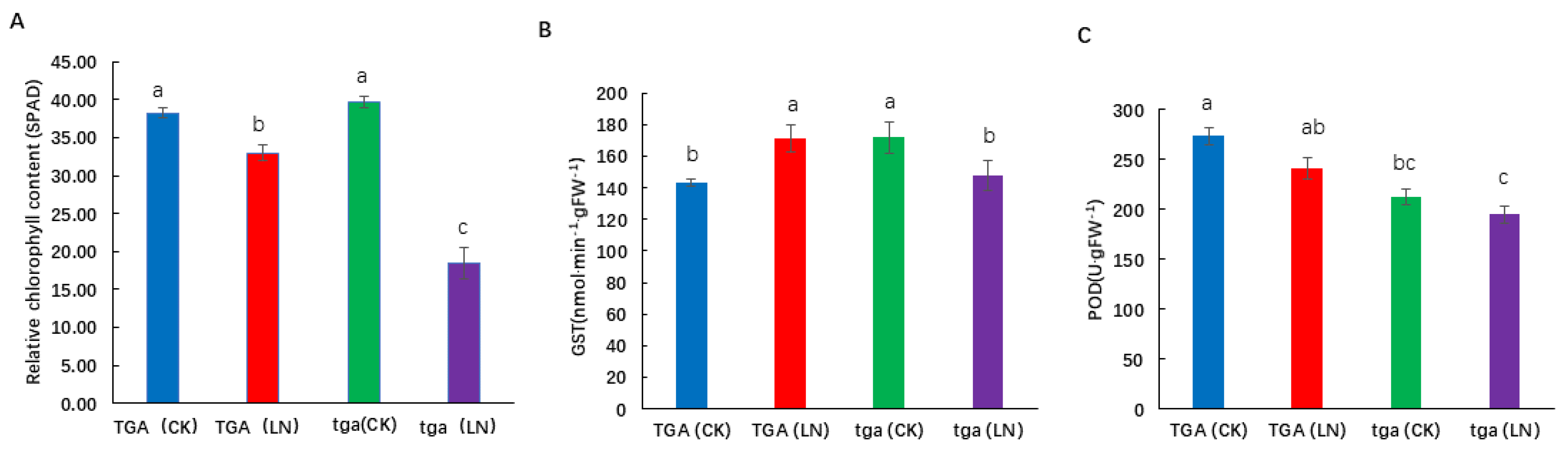
3.8. Phenotypic and Physiological Differences between Mutant tga and Wild-Type TGA in Hydroponic Treatment with Different Concentrations of Nitrate
4. Discussion
4.1. Phenotypic Differences in Response to Low Nitrogen Stress between Two Hybrids
4.2. DAPs Related to ‘Response to Stimulus’ in Two Maize Hybrids after Nitrogen Deficiency Treatment
4.3. DAPs Associated with Low Nitrogen Stress Tolerance
4.4. Possible Role of ZmTGA Gene in Maize Resistance to Low Nitrogen
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.N.; Xing, J.; Wang, Y.; Wen, H.T.; Wen, H.T.; Li, L.; Zhang, X.; Fan, Y.-L.; Zhao, J. Maize ABP2 enhances tolerance to drought and salt stress in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 2379–2393. [Google Scholar] [CrossRef] [Green Version]
- Gowik, U.; Westhoff, P. The path from C3 to C4 photosynthesis. Plant Physiol. 2011, 155, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosgey, J.R.; Moot, D.J.; Fletcher, A.L.; Mckenzie, B.A. Dry matter accumulation and post-silking N economy of ′stay-green′ maize (Zea mays L.) hybrids. Eur. J. Agron. 2013, 51, 43–52. [Google Scholar] [CrossRef]
- Rens, L.R.; Zotarelli, L.; Cantliffe, D.J.; Stoffella, P.J.; Gergela, D.; Fourman, D. Biomass Accumulation, Marketable Yield, and Quality of Atlantic Potato in Response to Nitrogen. Agron. J. 2015, 107, 931–942. [Google Scholar] [CrossRef]
- Pingle, G. Managing critical plant growth stages. Farmer’s Wkly. 2017, 2017, 46–49. [Google Scholar]
- Witte, C.P. Urea metabolism in plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef]
- Zhu, Q.; Liu, X.; Hao, T.; Zeng, M.; Shen, J.; Zhang, F.; de Vries, W. Crop land acidification increases risk of yield losses and food insecurity in China. Environ. Pollut. 2019, 256, 113145–113179. [Google Scholar] [CrossRef]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Vanessa, C.R.; Caas, R.A.; de la Torre, F.N.; Belen, P.M.; Avila, C.; Canovas, F.M. Molecular fundamentals of nitrogen uptake and transport in trees. J. Exp. Bot. 2017, 68, 2489–2500. [Google Scholar] [CrossRef]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Ma, X.J.; Cheng, Q.B.; Dou, P.; Yu, D.H.; Luo, Y.Q. Effects of nitrogen fertilizer on post-silking dry matter production and leaves function characteristics of low-nitrogen tolerance maize. Chin. J. Eco-Agric. 2016, 24, 17–26. [Google Scholar] [CrossRef]
- Ding, L.; Wang, K.J.; Jiang, G.M.; Biswas, D.K.; Xu, H.; Li, L.F.; Li, Y.H. Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Ann. Bot. 2015, 96, 925–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.J.; Gong, X.W.; Wang, H.L.; Dang, K.; Deng, X.P.; Feng, B.L. Low-nitrogen tolerance comprehensive evaluation and physiological response to nitrogen stress in broomcorn millet (Panicum miliaceum L.) seedling. Plant Physiol. Biochem. 2020, 151, 233–242. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.N.; Muhammad, K.; Xie, J.; Meng, X.P.; Han, Q.F.; Liu, T.N.; Han, J. Shoot and root traits of summer maize hybrid varieties with higher grain yields and higher nitrogen use efficiency at low nitrogen application rates. PeerJ 2019, 7, e7294. [Google Scholar] [CrossRef]
- Jakoby, M. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Kesarwani, M.; Jungmin, Y.; Dong, X. Genetic interaction of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007, 144, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.; Boden, E.; Arias, J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 2003, 15, 1846–1858. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Huh, S.U.; Kojima, M. The Cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [Green Version]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS protein SCL14 interacts with class Ⅱ TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef] [Green Version]
- Chern, W.; Singh, K.B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999, 19, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Frova, C. The plant glutathione transferase gene family: Genomic structure, function, expression and evolution. Physiol. Plant. 2003, 119, 469–479. [Google Scholar] [CrossRef]
- Köster, J.; Thurow, C.; Kruse, K.; Meier, A.; Iven, T.; Feussner, I.; Gatz, C. Xenobiotic-and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 2012, 159, 391–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-Lopez, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L.; et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef]
- Zhong, L.; Chen, D.; Min, D.; Li, W.; Xu, Z.; Zhou, Y.; Li, L. AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 457, 433–439. [Google Scholar] [CrossRef]
- Dong, A.Y.; Yang, Y.T.; Liu, S.T.; Zenda, T.; Liu, X.Y.; Wang, Y.F.; Li, L.; Chen, M.; Ma, Y. Comparative proteomics analysis of two maize hybrids revealed drought-stress tolerance mechanisms. Biotechnol. Biotechnol. Equip. 2020, 34, 763–780. [Google Scholar] [CrossRef]
- Du, W.; Xiong, C.W.; Ding, J.; Nybom, H.; Ruan, C.J.; Guo, H. TMT-based Quantitative Proteomics of Developing Sea Buckthorn Berries Reveals Candidate Proteins Related to Lipid Metabolism. J. Proteome Res. 2019, 18, 1958–1969. [Google Scholar] [CrossRef]
- Li, X.L.; Guo, L.G.; Zhou, B.Y.; Tang, X.M.; Chen, C.C.; Zhang, L.; Zhang, S.-Y.; Li, C.-F.; Xiao, K.; Dong, W.-X.; et al. Characterization of low-N responses in maize (Zea mays L.) cultivars with contrasting nitrogen use efficiency in the North China Plain. J. Integr. Agric. 2019, 18, 2141–2152. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lu, X.; Liu, J.; Ren, W.; Yang, Q.; Chai, Z.; Chen, R.; Wang, L.; Zhao, J.; Lang, Z.; Wang, H.; et al. Gene-Indexed Mutations in Maize. Mol. Plant 2018, 11, 496–504. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.G.; Wang, K.; Xu, C.; Zou, C.; Xie, C.X.; Xu, Y.B.; Li, W.X. Strand-specific RNA-Seq transcriptome analysis of genotypes with and without low-phosphorus tolerance provides novel insights into phosphorus-use efficiency in maize. BMC Plant Biol. 2016, 16, 222–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curci, P.L.; Cigliano, R.A.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 7, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Reddy, K.R.; Kakani, V.G.; Reddy, V.R. Nitrogen deficiency effects on plant growth, leaf photosynthesis, and hyperspectral reflectance properties of sorghum. Eur. J. Agron. 2005, 22, 391–403. [Google Scholar] [CrossRef]
- Makino, A.; Sakuma, H.; Sudo, E.; Mae, T. Differences between Maize and Rice in N-use Efficiency for Photosynthesis and Protein Allocation. Plant Cell Physiol. 2003, 44, 952–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takashima, T.; Hikosaka, K.; Hirose, T. Photosynthesis or persistence: Nitrogen allocation in leaves of evergreen and deciduous Quercus species. Plant Cell Environ. 2004, 27, 1047–1054. [Google Scholar] [CrossRef]
- Tazoe, Y.; Noguchi, K.O.; Terashima, I. Effects of growth light and nitrogen nutrition on the organization of the photosynthetic apparatus in leaves of a C4 plant, Amaranthus cruentus. Plant Cell Environ. 2005, 29, 691–700. [Google Scholar] [CrossRef]
- Hou, P.; Gao, Q.; Xie, R.Z.; Li, S.K.; Meng, Q.F.; Ernest, A.K.; Römheld, V.; Müller, T.; Zhang, F.; Cui, Z.; et al. Grain yields in relation to N requirement: Optimizing nitrogen management for spring maize grown in China. Field Crops Res. 2012, 129, 1–6. [Google Scholar] [CrossRef]
- Sultana, N.; Islam, S.; Juhasz, A.; Yang, R.C.; She, M.Y.; Alhabbar, Z.; Zhang, J.; Ma, W. Transcriptomic Study for Identification of Major Nitrogen Stress Responsive Genes in Australian Bread Wheat Hybrids. Front. Genet. 2020, 11, 1086. [Google Scholar] [CrossRef]
- Huang, Y.P.; Huang, Y.W.; Chen, I.H.; Denby, K. Plasma membrane-associated cation-binding protein 1-like protein negatively regulates intercellular movement of BaMV. J. Exp. Bot. 2017, 68, 4765–4774. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Tang, N.; Ma, S.; Zong, W.; Li, J.; Li, X.; Xiang, Y.; Guo, Z.; Li, J.; Li, X.; Xiang, Y.; et al. MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 2016, 28, 2161–2177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Hu, Q.; Zhou, M.; Vandenbrink, J.; Li, D.; Menchyk, N.; Reighard, S.; Norris, A.; Liu, H.; Sun, D.; et al. Heterologous expression of OsSIZ1, a rice SUMO E3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass. Plant Biotechnol. J. 2013, 11, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Lin, S.I. Molecular regulators of phosphate homeostasis in plants. J. Exp Bot. 2009, 60, 1427–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef] [Green Version]
- Peng, M.; Hannam, C.; Gu, H.; Bi, Y.M.; Rothstein, S.J. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2010, 50, 320–337. [Google Scholar] [CrossRef]
- Negi, S.; Barry, A.N.; Friedland, N.; Sudasinghe, N.; Sayre, R. Impact of nitrogen limitation on biomass, photo-synthesis, and lipid accumulation in chlorella sorokiniana. J. Appl. Phycol. 2015, 28, 803–812. [Google Scholar] [CrossRef]
- Jing, X.Q.; Zhou, M.R.; Nie, X.M.; Zhang, L.; Chen, K.M. Osgstu6 contributes to cadmium stress tolerance in rice by involving in intracellular ROS homeostasis. J. Plant Growth Regul. 2020, 40, 945–961. [Google Scholar] [CrossRef]
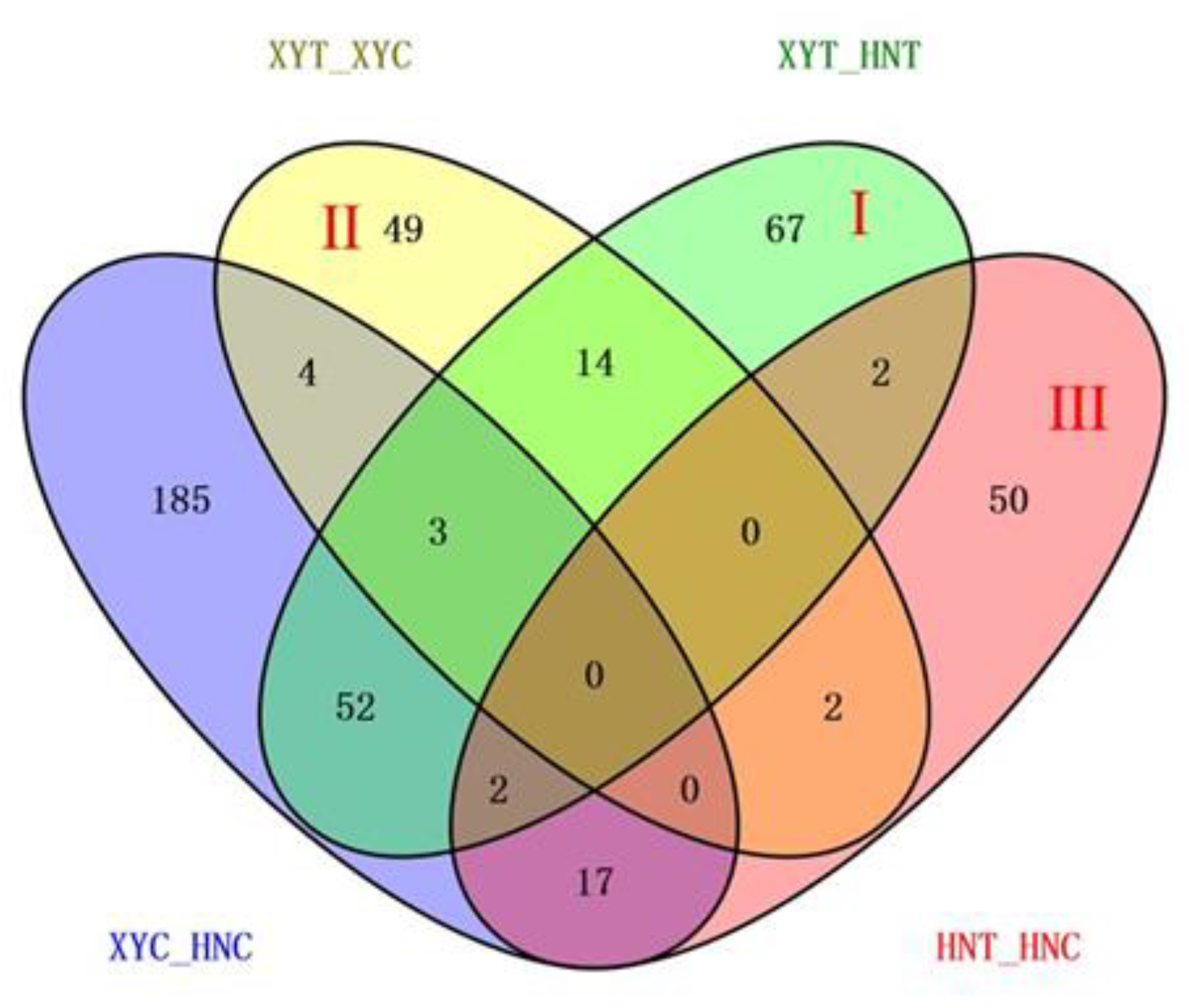


| Comparisons * | Upregulated | Downregulated | Total |
|---|---|---|---|
| HNT_HNC | 26 | 47 | 73 |
| XYT_XYC | 46 | 26 | 72 |
| XYT_HNT | 80 | 60 | 140 |
| XYC_HNC | 155 | 108 | 263 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, N.; Liu, S.; Dong, A.; Zenda, T.; Liu, X.; Li, J.; Duan, H. Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene. Genes 2022, 13, 670. https://doi.org/10.3390/genes13040670
Wang Y, Wang N, Liu S, Dong A, Zenda T, Liu X, Li J, Duan H. Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene. Genes. 2022; 13(4):670. https://doi.org/10.3390/genes13040670
Chicago/Turabian StyleWang, Yafei, Nan Wang, Songtao Liu, Anyi Dong, Tinashe Zenda, Xinyue Liu, Jiao Li, and Huijun Duan. 2022. "Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene" Genes 13, no. 4: 670. https://doi.org/10.3390/genes13040670
APA StyleWang, Y., Wang, N., Liu, S., Dong, A., Zenda, T., Liu, X., Li, J., & Duan, H. (2022). Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene. Genes, 13(4), 670. https://doi.org/10.3390/genes13040670






