Bioinformatics Analysis Highlight Differentially Expressed CCNB1 and PLK1 Genes as Potential Anti-Breast Cancer Drug Targets and Prognostic Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
- Building a DGElist object, and then filtering out the low expression genes using the FilterbyExpr function attached to the edgeR toolkit;
- For the convenience of analysis, the expression levels of different genes in the database have been assigned. The genes’ average expression levels bigger than 1 in each sample were screened;
- The expressed genes were screened in 75% of the samples.
2.2. Identification of Differentially Expressed Genes
2.3. Weighted Genes Correlation Network Analysis
2.4. Construction of Protein-Protein Interaction Network
2.5. Screening of Core Modules and Hub Genes
2.6. Expression Analysis
2.7. Survival Analysis
2.8. Kaplan-Meier Plotter
2.9. Analysis of Candidate Targets
2.10. Gene Enrichment Analysis
- Gene Ontology (GO) and KEGG enriched module genes obtained by WGCNA;
- DAVID database was used to conduct GO and KEGG enrichment analysis on the modules with the highest scores screened by the plug-in MCODE;
- GEPIA 2 database was used to screen ten genes that similar to each hub gene’s expression pattern. The Metascape (https://metascape.org/, accessed on 6 March 2022) was chosen for enrichment analysis.
2.11. Tools
3. Results
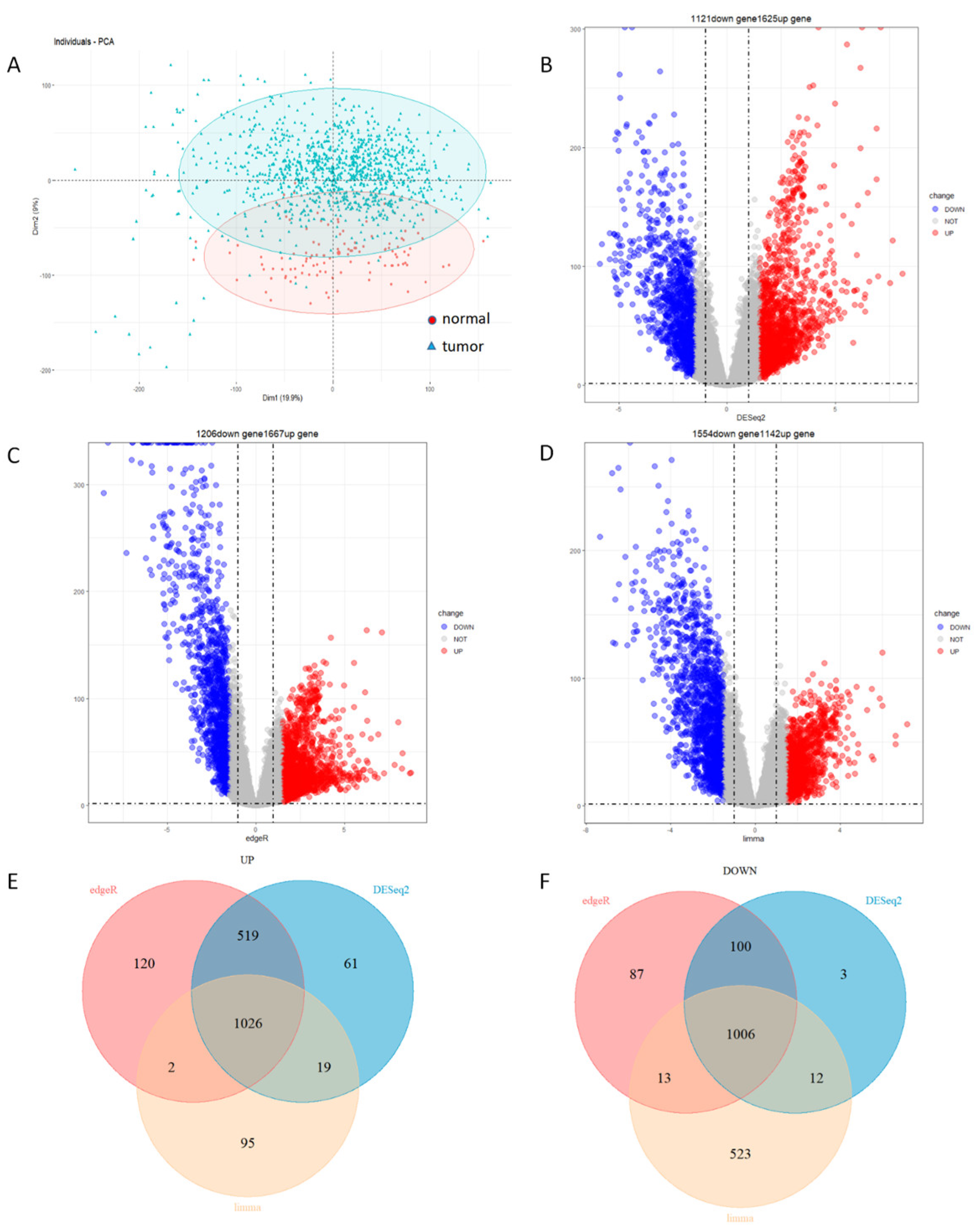
3.1. Identification of 1026 Up-Regulated Genes and 1006 Down-Regulated Genes
3.2. Identification of 693 Module Genes Correlated with Breast Cancer
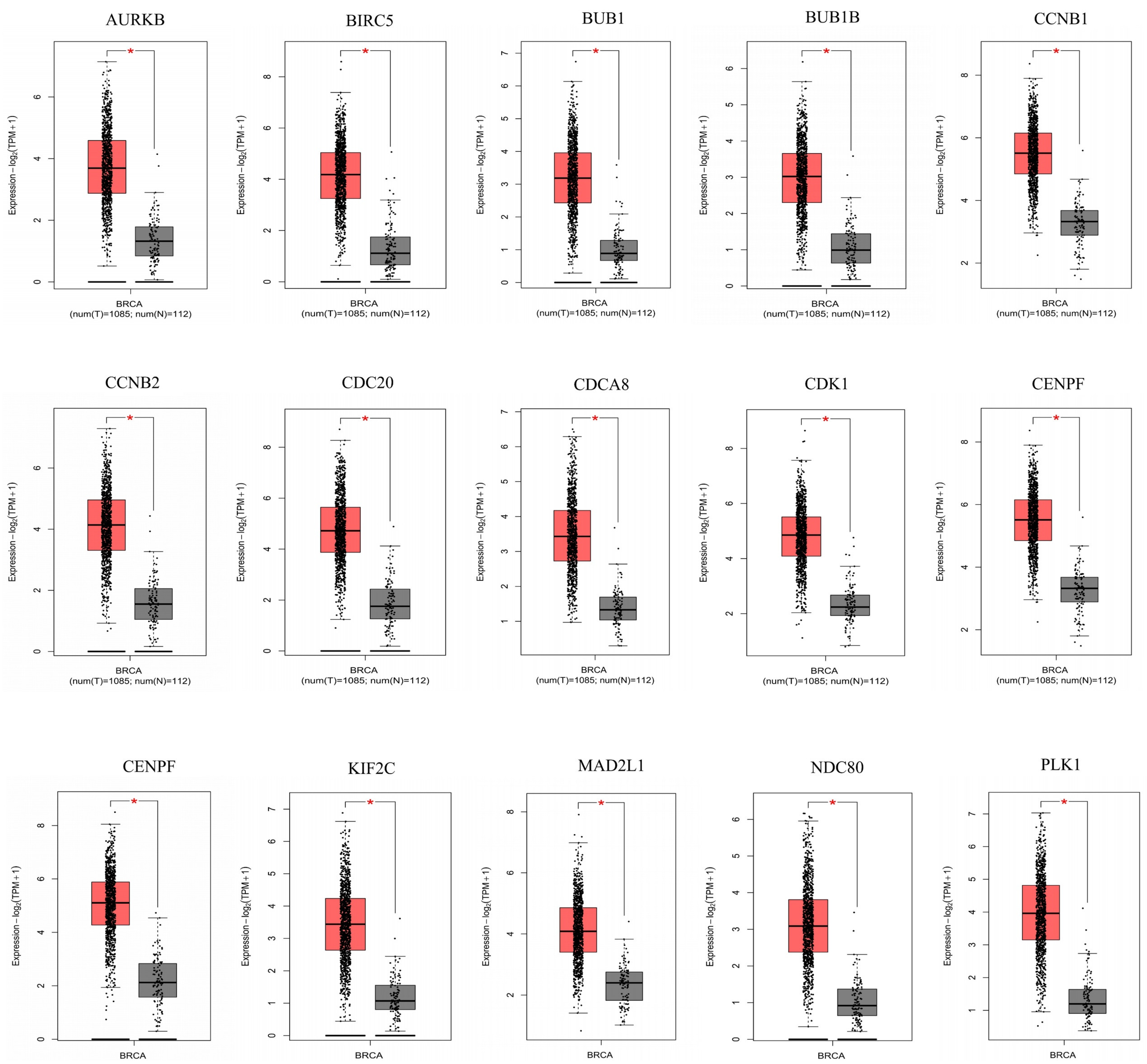
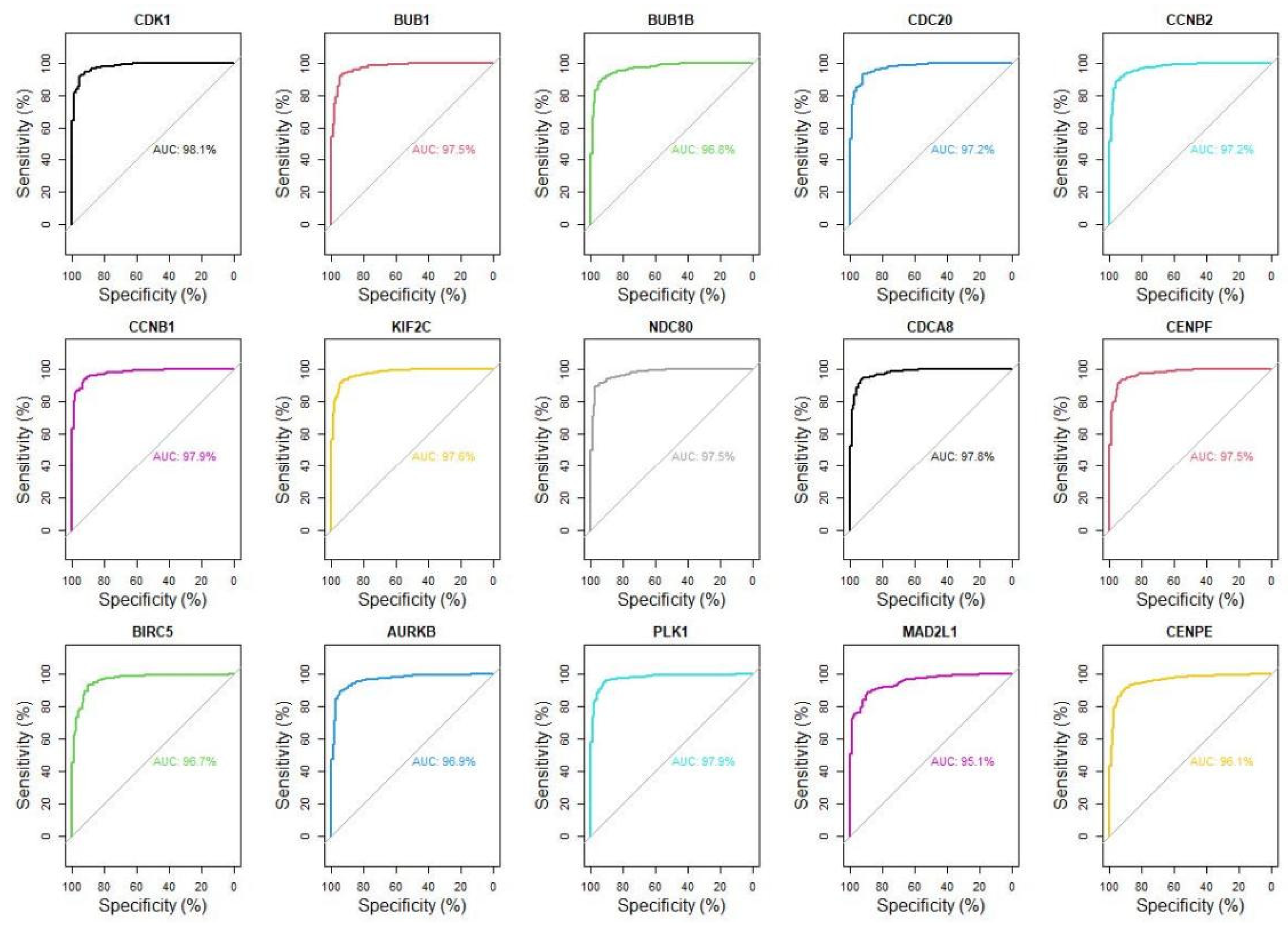
3.3. A Total of Fifteen Hub Genes Were Selected by the PPI Network
3.4. Fifteen Hub Genes Were Highlighted as Potential Therapeutic Drug Targets
3.5. Highly Expressed CCNB1 and PLK1 in Breast Cancer Were Associated with a Low Survival Rate
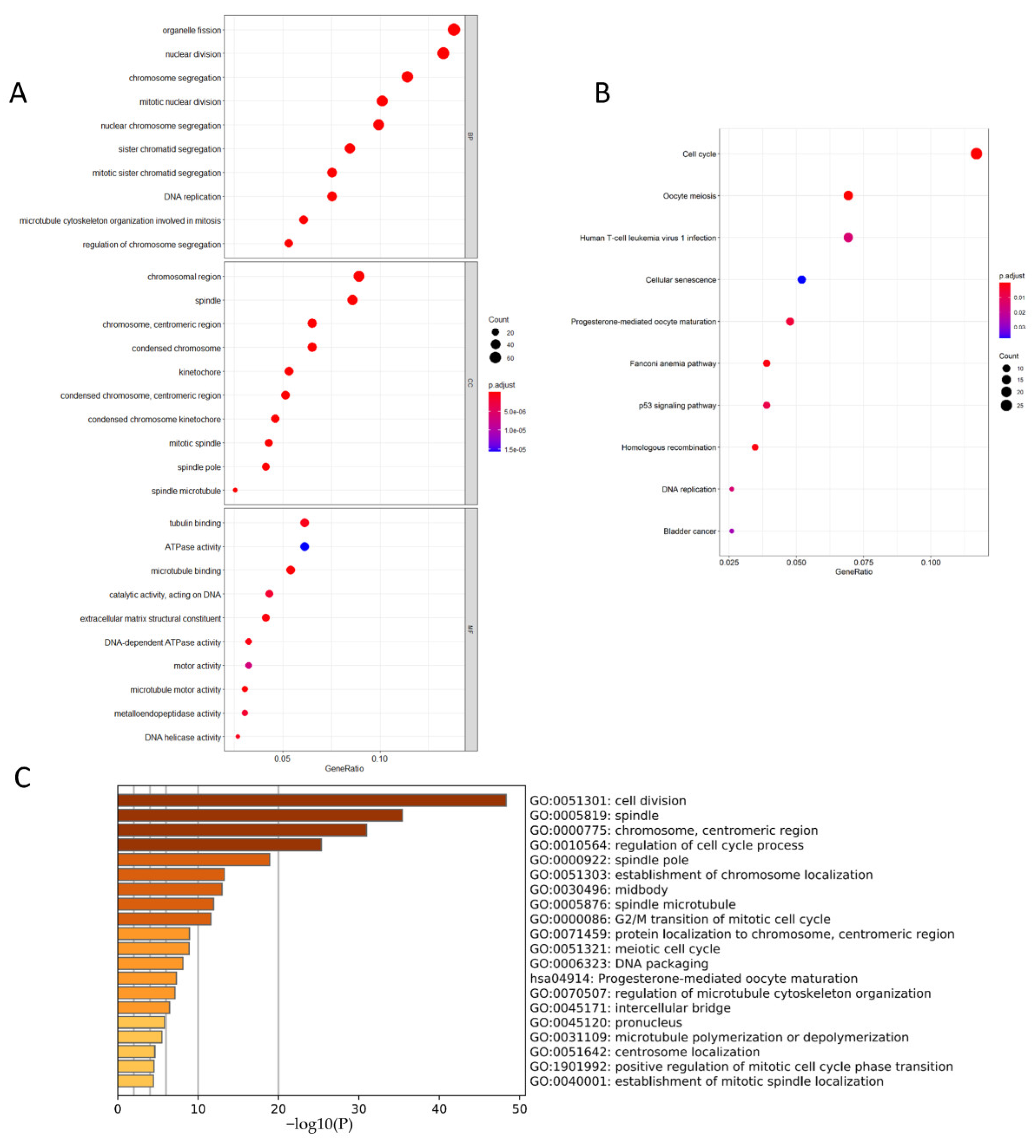
3.6. CCNB1 and PLK1 Were Enriched in Breast Cancer Cell Division and Cell Cycle-Related Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, L.; Strasser-Weippl, K.; Li, J.-J.; St. Louis, J.; Finkelstein, D.M.; Yu, K.-D.; Chen, W.-Q.; Shao, Z.-M.; Gross, P.E. Breast cancer in China. Lancet Oncol. 2014, 15, 279–289. [Google Scholar] [CrossRef]
- Siegel, L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Latest Global Cancer Data: Cancer Burden Rises to 19.3 Million New Cases and 10.0 Million Cancer Deaths in 2020 Questions and Answers (Q&A). 2020. Available online: https://www.iarc.who.int/faq/latest-global-cancer-data-2020-qa/ (accessed on 6 March 2022).
- Ebata, A.; Suzuki, T.; Takagi, K.; Miki, Y.; Onodera, Y.; Nakamura, Y.; Fujishima, F.; Ishida, K.; Watanabe, M.; Tamaki, K.; et al. Oestrogen-induced genes in ductal carcinoma in situ: Their comparison with invasive ductal carcinoma. Endocr. Related Cancer 2012, 19, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Khaled, W.; Lee, S.C.; Stingl, J.; Chen, X.; Ali, H.R.; Rueda, O.M.; Hadi, F.; Wang, J.; Yu, Y.; Chin, S.-F.; et al. BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat. Commun. 2015, 6, 5987. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Stewart, S.; Nagtegaal, I.; Luo, J.; Wu, Y.; Colditz, G.; Medina, D.; Allred, D.C. Differentially Expressed Genes Regulating the Progression of Ductal Carcinoma In Situ to Invasive Breast Cancer. Cancer Res. 2012, 72, 4574–4586. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Tian, J. FOXK1, Regulated by miR-365-3p, Promotes Cell Growth and EMT Indicates Unfavorable Prognosis in Breast Cancer. Onco. Targets Ther. 2020, 13, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Hisamatsu, Y.; Tokunaga, E.; Yamashita, N.; Akiyoshi, S.; Okada, S.; Nakashima, Y.; Taketani, K.; Aishima, S.; Oda, Y.; Morita, M.; et al. Impact of GATA-3 and FOXA1 expression in patients with hormone recep-tor-positive/HER2-negative breast cancer. Breast Cancer 2015, 22, 520–528. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Lee, C.-F.; Lai, H.-T.; Yu, C.-T.; Lee, J.-E.; Zuo, H.; Tsai, S.Y.; Tsai, M.-J.; Ge, K.; Wan, Y.; et al. Opposing Functions of BRD4 Isoforms in Breast Cancer. Mol. Cell 2020, 78, 1114–1132.e10. [Google Scholar] [CrossRef]
- Hinz, N.; Jücker, M. Distinct functions of AKT isoforms in breast cancer: A comprehensive review. Cell Commun. Signal. 2019, 17, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Han, B.; Chen, J.; Wiedemeyer, R.; Orsulic, S.; Bose, S.; Zhang, X.; Karlan, B.Y.; Giuliano, A.E.; Cui, Y.; et al. FOXC1 is a critical mediator of EGFR function in human basal-like breast cancer. Ann. Surg. Oncol. 2014, 21, S758–S766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Fan, X.; Hao, M.; Wang, J.; Zhou, X.; Sun, X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol. Cancer 2016, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Zhu, L.; Li, L.; Li, J.; Xu, L.; Feng, J.; Liu, Y. Digital Gene Expression Analysis Provides Insight into the Transcript Profile of the Genes Involved in Aporphine Alkaloid Biosynthesis in Lotus (Nelumbo nucifera). Front. Plant Sci. 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Stupnikov, A.; McInerney, C.; Savage, K.; McIntosh, S.; Emmert-Streib, F.; Kennedy, R.; Salto-Tellez, M.; Prise, K.; McArt, D. Robustness of differential gene expression analysis of RNA-seq. Comput. Struct. Biotechnol. J. 2021, 19, 3470–3481. [Google Scholar] [CrossRef]
- Tani, J.; Faustine; Sufian, J.T. Updates on current advances in gene therapy. West Indian Med. J. 2011, 60, 188–194. [Google Scholar]
- Zhang, H.; Zhang, X.; Li, X.; Meng, W.; Bai, Z.; Rui, S.; Wang, Z.; Zhou, W.; Jin, X. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J. Cell. Physiol. 2018, 234, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Androic, I.; Krämer, A.; Yan, R.; Rödel, F.; Gätje, R.; Kaufmann, M.; Strebhardt, K.; Yuan, J. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer 2008, 8, 391. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Du, W.W.; Lyu, J.; Dong, J.; Zhang, C.; Yang, W.; He, A.; Kwok, Y.S.S.; Ma, J.; Wu, N.; et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 2018, 25, 2195–2208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niméus-Malmström, E.; Koliadi, A.; Ahlin, C.; Holmqvist, M.; Holmberg, L.; Amini, R.-M.; Jirström, K.; Wärnberg, F.; Blomqvist, C.; Fernö, M.; et al. Cyclin B1 is a prognostic proliferation marker with a high reproducibility in a population-based lymph node negative breast cancer cohort. Int. J. Cancer 2009, 127, 961–967. [Google Scholar] [CrossRef]
- Glover, D.M.; Hagan, I.M.; Tavares, A.A. olo-like kinases: A team that plays throughout mitosis. Genes Dev. 1998, 12, 3777–3787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, B.T.; Strebhardt, K. Polo-Like Kinase 1: Target and Regulator of Transcriptional Control. Cell Cycle 2006, 5, 2881–2885. [Google Scholar] [CrossRef] [Green Version]
- Spänkuch, B.; Heim, S.; Kurunci-Csacsko, E.; Lindenau, C.; Yuan, J.; Kaufmann, M.; Strebhardt, K. Down-regulation of Polo-like Kinase 1 Elevates Drug Sensitivity of Breast Cancer Cells In vitro and In vivo. Cancer Res. 2006, 66, 5836–5846. [Google Scholar] [CrossRef] [Green Version]
- Wierer, M.; Verde, G.; Pisano, P.; Molina, H.; Font-Mateu, J.; Di Croce, L.; Beato, M. PLK1 Signaling in Breast Cancer Cells Cooperates with Estrogen Receptor-Dependent Gene Transcription. Cell Rep. 2013, 3, 2021–2032. [Google Scholar] [CrossRef] [Green Version]
- Izadi, S.; Nikkhoo, A.; Farsangi, M.H.; Namdar, A.; Azizi, G.; Mohammadi, H.; Yousefi, M.; Jadidi-Niaragh, F. CDK1 in Breast Cancer: Implications for Theranostic Potential. Anti-Cancer Agents Med. Chem. 2020, 20, 758–767. [Google Scholar] [CrossRef]
- Cheng, S.; Castillo, V.; Sliva, D. CDC20 associated with cancer metastasis and novel mushroom-derived CDC20 inhibitors with antimetastatic activity. Int. J. Oncol. 2019, 54, 2250–2256. [Google Scholar] [CrossRef]
- Wu, S.; Su, R.; Jia, H. Cyclin B2 (CCNB2) Stimulates the Proliferation of Triple-Negative Breast Cancer (TNBC) Cells In Vitro and In Vivo. Dis. Markers 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Zhu, L.; Pan, Y.; Chen, X.; Hou, P. BUB1 promotes proliferation of liver cancer cells by activating SMAD2 phosphorylation. Oncol. Lett. 2020, 19, 3506–3512. [Google Scholar] [CrossRef]
- Koyuncu, D.; Sharma, U.; Goka, E.T.; Lippman, M.E. Spindle assembly checkpoint gene BUB1B is essential in breast cancer cell survival. Breast Cancer Res. Treat. 2020, 185, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-F.; Xie, Y.-X.; Qian, Y.-C.; Wang, M.; Liu, L.-Z.; Shu, Y.-Q.; Bai, X.-M.; Jiang, B.-H. TBX15/miR-152/KIF2C pathway regulates breast cancer doxorubicin resistance via promoting PKM2 ubiquitination. Cancer Cell Int. 2021, 21, 542. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Huang, L.; Liu, L.; Qin, H.; Song, Z. Nuclear division cycle 80 promotes malignant progression and predicts clinical outcome in colorectal cancer. Cancer Med. 2018, 7, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wen, X.; He, H.; Zheng, L.; Yang, Y.; Yang, J.; Liu, H.; Zhou, X.; Yang, C.; Chen, Y.; et al. Knockdown of CDCA8 inhibits the proliferation and enhances the apoptosis of bladder cancer cells. PeerJ 2020, 8, e9078. [Google Scholar] [CrossRef]
- Shahid, M.; Lee, M.Y.; Piplani, H.; Andres, A.M.; Zhou, B.; Yeon, A.; Kim, M.; Kim, H.L.; Kim, J. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metab-olism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle 2018, 17, 2802–2818. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-Y.; Chan, H.-H.; Chen, S.-H.; Sarvagalla, S.; Chen, P.-S.; Coumar, M.S.; Cheng, S.M.; Chang, Y.-C.; Lin, C.-H.; Leung, E.; et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy 2020, 16, 1296–1313. [Google Scholar] [CrossRef] [Green Version]
- Nie, M.; Wang, Y.; Yu, Z.; Li, X.; Deng, Y.; Wang, Y.; Yang, D.; Li, Q.; Zeng, X.; Ju, J.; et al. AURKB promotes gastric cancer progression via activation of CCND1 expression. Aging 2020, 12, 1304–1321. [Google Scholar] [CrossRef]
- Li, Y.; Benezra, R. Identification of a Human Mitotic Checkpoint Gene: hsMAD2. Science 1996, 274, 246–248. [Google Scholar] [CrossRef]
- Shan, L.; Zhao, M.; Lu, Y.; Ning, H.; Yang, S.; Song, Y.; Chai, W.; Shi, X. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int. J. Oncol. 2019, 55, 257–266. [Google Scholar]
- Berns, K.; Horlings, H.M.; Hennessy, B.T.; Madiredjo, M.; Hijmans, E.M.; Beelen, K.; Linn, S.C.; Gonzalez-Angulo, A.M.; Stemke-Hale, K.; Hauptmann, M.; et al. A Functional Genetic Approach Identifies the PI3K Pathway as a Major Determinant of Trastuzumab Resistance in Breast Cancer. Cancer Cell 2007, 12, 395–402. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, L.; Liu, Q.; Cui, H.; Zheng, Y.; Wu, C. Bioinformatics Analysis Highlight Differentially Expressed CCNB1 and PLK1 Genes as Potential Anti-Breast Cancer Drug Targets and Prognostic Markers. Genes 2022, 13, 654. https://doi.org/10.3390/genes13040654
Fang L, Liu Q, Cui H, Zheng Y, Wu C. Bioinformatics Analysis Highlight Differentially Expressed CCNB1 and PLK1 Genes as Potential Anti-Breast Cancer Drug Targets and Prognostic Markers. Genes. 2022; 13(4):654. https://doi.org/10.3390/genes13040654
Chicago/Turabian StyleFang, Leiming, Qi Liu, Hongtu Cui, Yunji Zheng, and Chengjun Wu. 2022. "Bioinformatics Analysis Highlight Differentially Expressed CCNB1 and PLK1 Genes as Potential Anti-Breast Cancer Drug Targets and Prognostic Markers" Genes 13, no. 4: 654. https://doi.org/10.3390/genes13040654
APA StyleFang, L., Liu, Q., Cui, H., Zheng, Y., & Wu, C. (2022). Bioinformatics Analysis Highlight Differentially Expressed CCNB1 and PLK1 Genes as Potential Anti-Breast Cancer Drug Targets and Prognostic Markers. Genes, 13(4), 654. https://doi.org/10.3390/genes13040654




