Global Screening and Functional Identification of Major HSPs Involved in PVY Infection in Potato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the DnaJ Genes in Potato
2.2. Sequence Analysis and Classification of Potato DnaJ Genes
2.3. Phylogenetic Analysis of Potato DnaJ Genes
2.4. Chromosomal Distribution and Gene Duplication Events
2.5. Plant Materials
2.6. PVY Inoculation
2.7. RNA-Seq Data Analysis
2.8. Virus-Induced Gene Silencing
2.9. RNA Isolation, qRT-PCR, and ELISA Analysis
3. Results
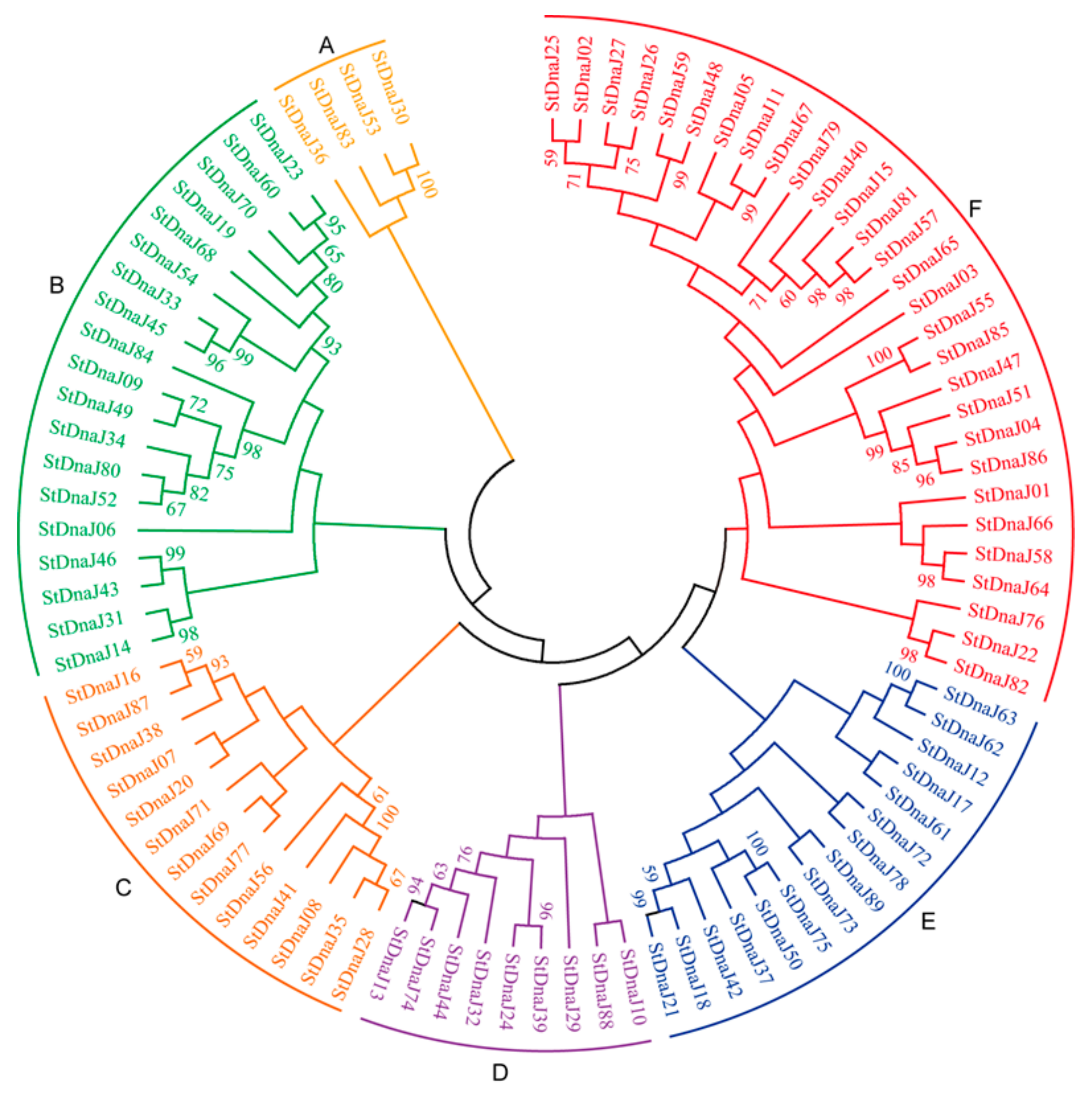
3.1. Identification, Classification, and Phylogenetic Analysis of Potato DnaJ Genes
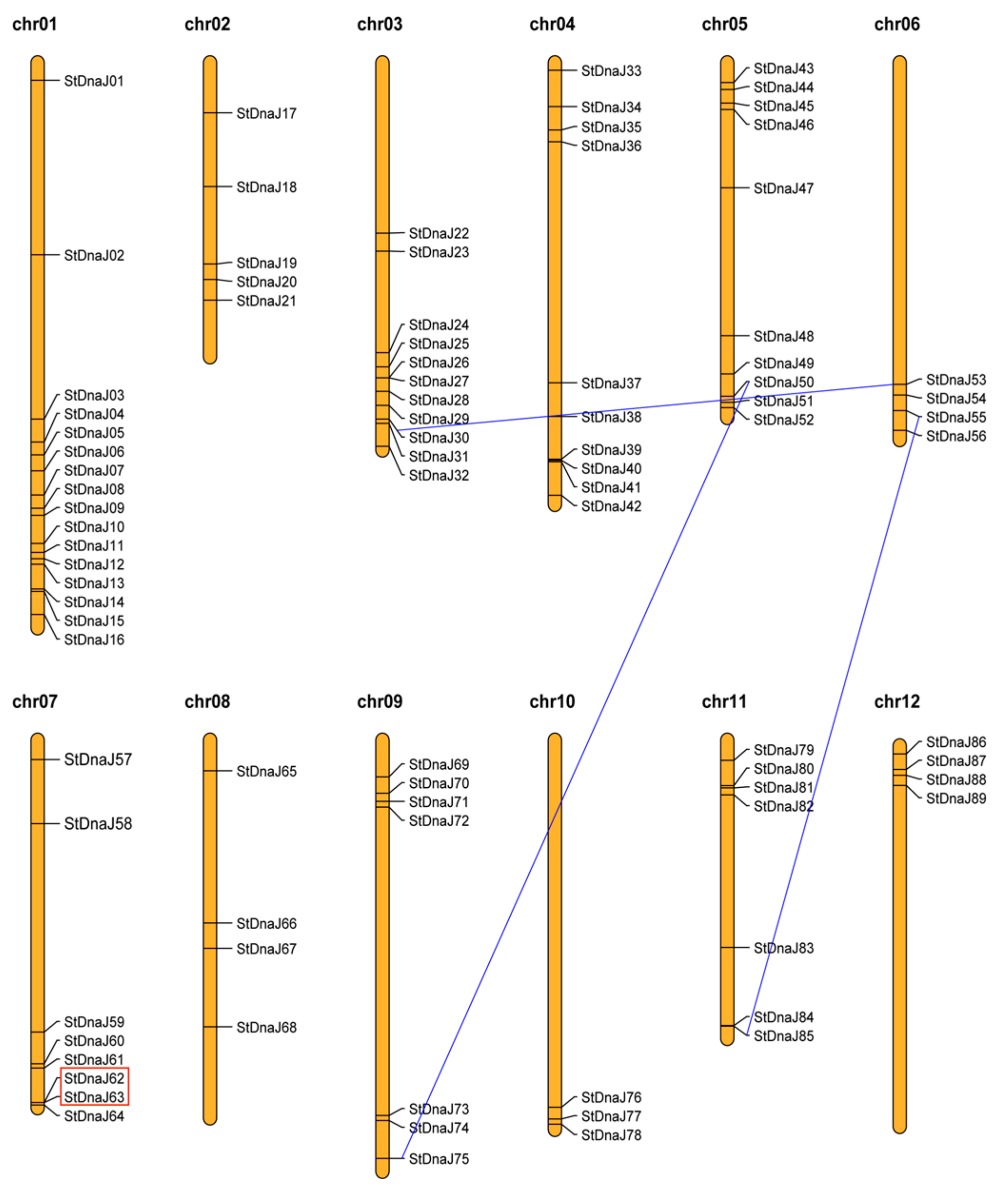
3.2. Chromosomal Locations and Gene Duplication Events of the StDnaJ Family
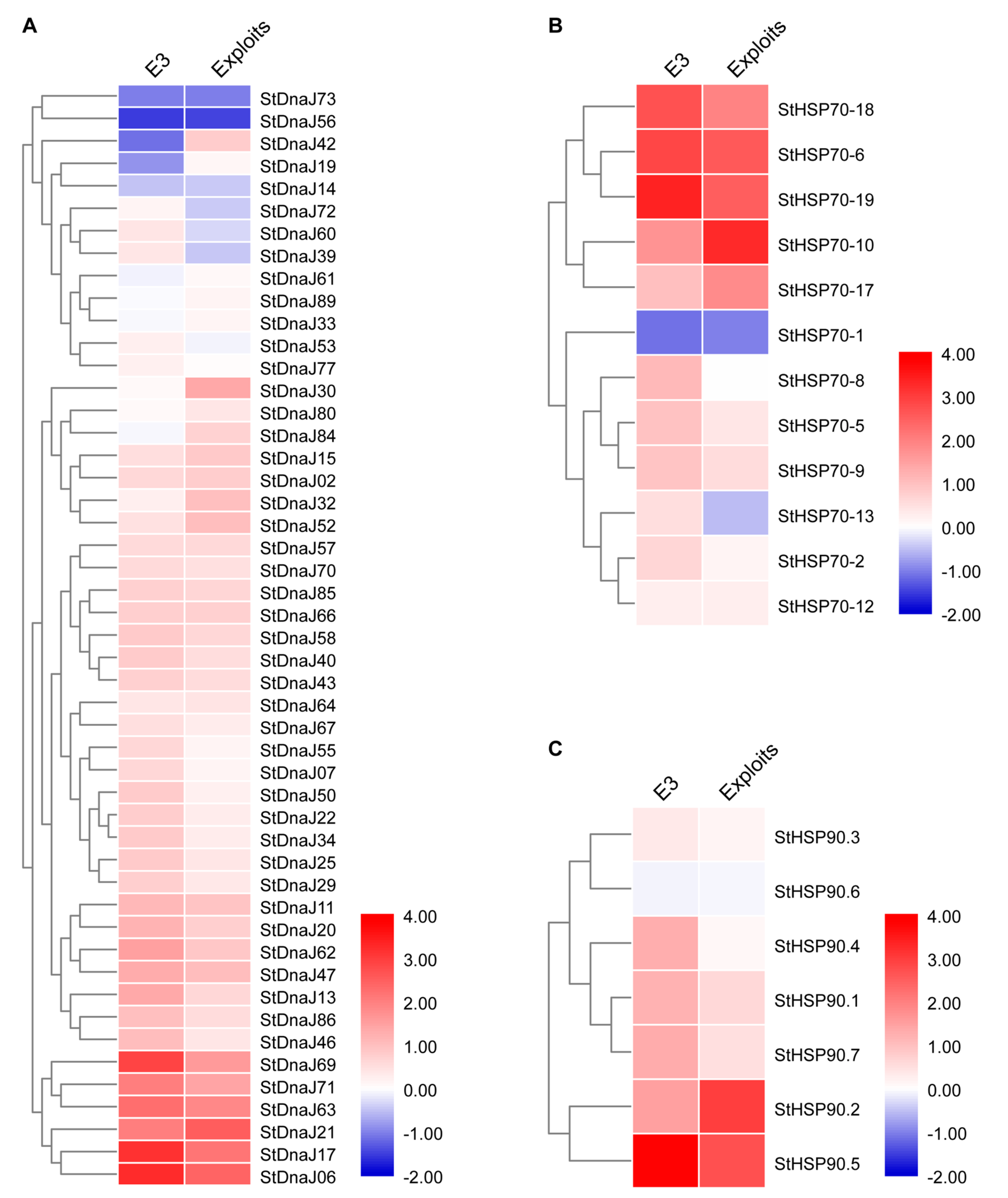
3.3. Expression Profiles of StDnaJ, StHSP70, and StHSP90 Families under PVY Infection
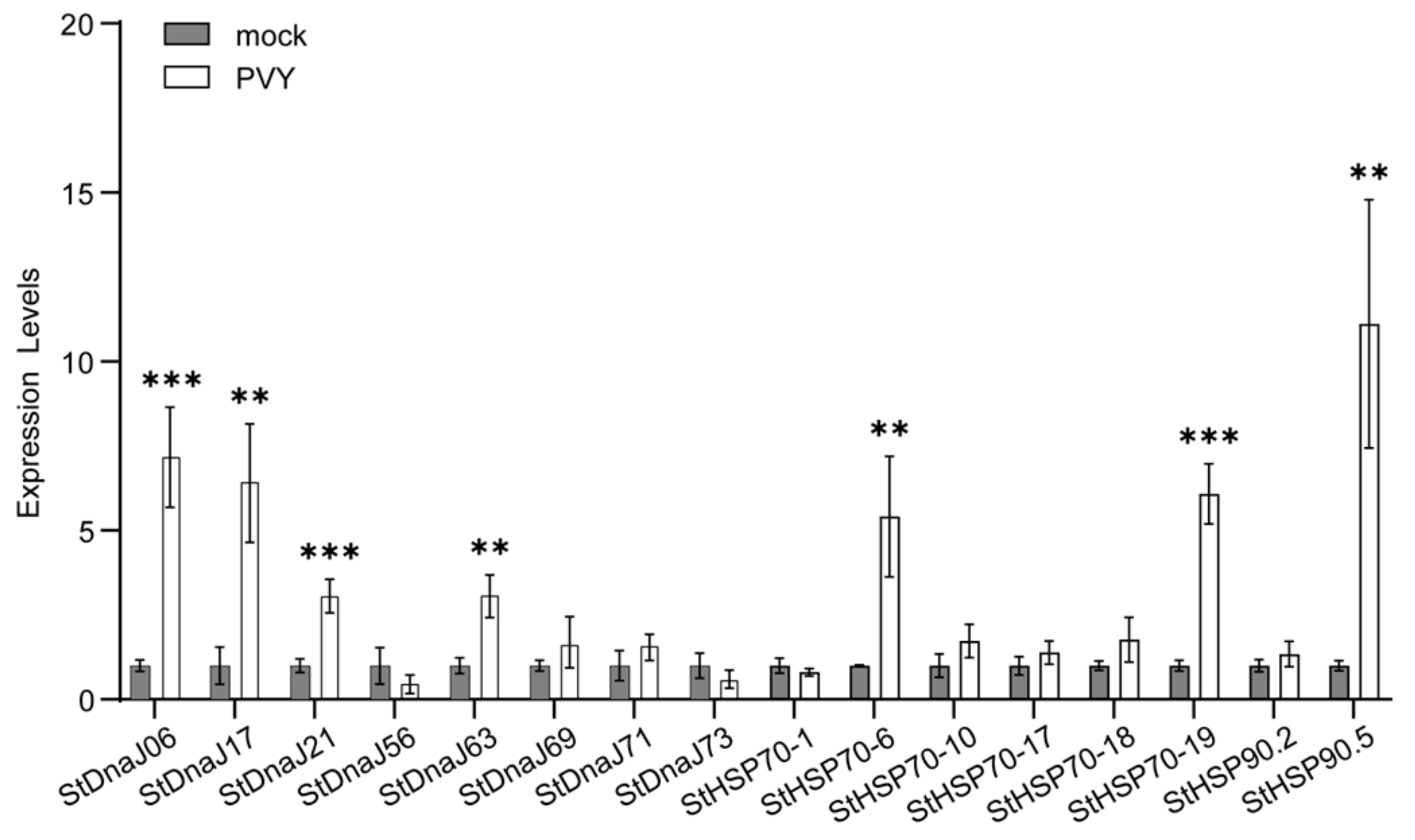
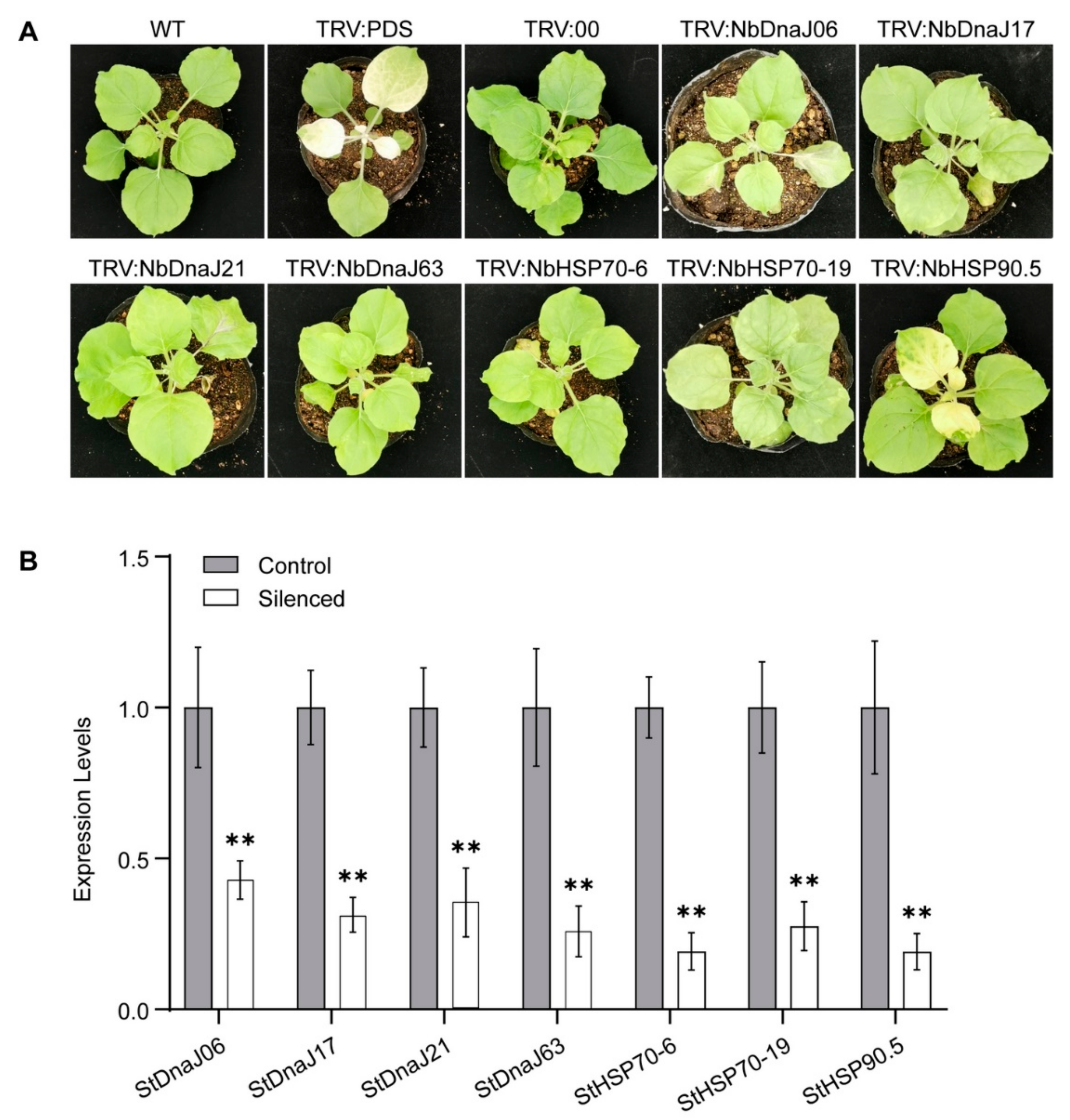
3.4. Virus-Induced Gene Silencing (VIGS) Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scott, G.J.; Rosegrant, M.W.; Ringler, C. Global projections for root and tuber crops to the year 2020. Food Policy 2000, 25, 561–597. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactive Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection with Potato Virus Y. Front. Microbiol. 2018, 9, 2582. [Google Scholar] [PubMed] [Green Version]
- Radcliffe, E.B.; Ragsdale, D.W. Aphid-transmitted potato viruses: The importance of understanding vector biology. Am. J. Potato Res. 2002, 79, 353–386. [Google Scholar] [CrossRef]
- Carey, C.C.; Gorman, K.F.; Rutherford, S. Modularity and intrinsic evolvability of Hsp90-buffered change. PLoS ONE 2006, 1, e76. [Google Scholar] [CrossRef]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sharma, A.; Mishra, M.; Mishra, R.K.; Chowdhuri, D.K. Heat shock proteins in toxicology: How close and how far? Life Sci. 2010, 86, 377–384. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Pulido, P.; Leister, D. Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytol. 2018, 217, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Caplan, A.J.; Cyr, D.M.; Douglas, M.G. Eukaryotic Homologs of Escherichia-Coli Dnaj—A Diverse Protein Family That Functions with Hsp70 Stress Proteins. Mol. Biol. Cell. 1993, 4, 555–563. [Google Scholar] [CrossRef] [Green Version]
- Silver, P.A.; Way, J.C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 1993, 74, 5–6. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Pearl, L.H.; Prodromou, C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006, 75, 271–294. [Google Scholar] [CrossRef]
- Zhao, R.; Houry, W.A. Molecular interaction network of the Hsp90 chaperone system. In Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks, 2nd ed.; Csermely, P., Vígh, L., Eds.; Springer: New York, NY, USA, 2007; Volume 594, pp. 27–36. [Google Scholar]
- Kozieł, E.; Surowiecki, P.; Przewodowska, A.; Bujarski, J.J.; Otulak-Kozieł, K. Modulation of Expression of PVYNTN RNA-Dependent RNA Polymerase (NIb) and Heat Shock Cognate Host Protein HSC70 in Susceptible and Hypersensitive Potato Cultivars. Vaccines 2021, 9, 1254. [Google Scholar] [CrossRef]
- Hýsková, V.; Bělonožníková, K.; Doričová, V.; Kavan, D.; Gillarová, S.; Henke, S.; Ryšlavá, H.; Čeřovská, N. Effects of heat treatment on metabolism of tobacco plants infected with Potato virus Y. Plant Biol. 2021, 23, 131–141. [Google Scholar] [CrossRef]
- Shimizu, T.; Yoshii, A.; Sakurai, K.; Hamada, K.; Yamaji, Y.; Suzuki, M.; Namba, S.; Hibi, T. Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of tobacco mosaic virus. Arch. Virol. 2009, 154, 959–967. [Google Scholar] [CrossRef]
- Cho, S.Y.; Cho, W.K.; Sohn, S.H.; Kim, K.H. Interaction of the host protein NbDnaJ with Potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 2012, 417, 451–456. [Google Scholar] [CrossRef]
- Hafren, A.; Hofius, D.; Ronnholm, G.; Sonnewald, U.; Makinen, K. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 2010, 22, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Lõhmus, A.; Hafrén, A.; Mäkinen, K. Coat protein regulation by CK2, CPIP, HSP70, and CHIP is required for potato virus A replication and coat protein accumulation. J. Virol. 2017, 91, e01316-16. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J. Virol. 2012, 86, 12091–12104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, L.; Zhao, J.; Du, Y.; Zhao, X.; Han, M.; Liu, Y. Hsp90 Interacts With Tm-22 and Is Essential for Tm-22-Mediated Resistance to Tobacco mosaic virus. Front. Plant Sci. 2018, 9, 411. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Pang, X.; Cheng, Y.; Yin, Y.; Zhang, Q.; Su, W.; Hu, B.; Guo, Q.; Ha, S.; Zhang, J.; et al. The Hsp70 Gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep. 2018, 8, 16628. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Y.; Ye, M.; Wang, D.; Chen, Q. Evolutionary history of the heat shock protein 90 (Hsp90) family of 43 plants and characterization of Hsp90s in Solanum tuberosum. Mol. Biol. Rep. 2020, 47, 6679–6691. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, X.; Liang, Z.; Nie, B.; Murphy, A.; Singh, A. Studies on varietal response to different strains of Potato virus Y (PVY) reveal hypersensitive resistance in Exploits to PVYO and extreme resistance in F87084 to all tested strains. Am. J. Potato Res. 2015, 92, 23–31. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Chakravarthy, S.; Martin, G.B. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 2009, 28, e1292. [Google Scholar]
- Nie, B.; Singh, M.; Sullivan, A.; Singh, R.P.; Xie, C.; Nie, X. Recognition and Molecular Discrimination of Severe and Mild PVYO Variants of Potato virus Y in Potato in New Brunswick, Canada. Plant Dis. 2011, 95, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.P.; McLaren, D.L.; Nie, X.; Singh, M. Possible Escape of a Recombinant Isolate of Potato virus Y by Serological Indexing and Methods of its Detection. Plant Dis. 2003, 87, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Brigneti, G.; Martin-Hernandez, A.M.; Jin, H.; Chen, J.; Baulcombe, D.C.; Baker, B.; Jones, J.D. Virus-induced gene silencing in Solanum species. Plant J. 2004, 39, 264–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Walsh, P.; Bursac, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Nie, B.; Tu, Z.; Li, C.; Murphy, A.; Singh, M.; Song, B.; Zhang, S.; Xie, C.; Nie, X. Extreme Resistance to Potato Virus A in Potato Cultivar Barbara is Independently Mediated by Ra and Rysto. Plant Dis. 2021, 105, 3344–3348. [Google Scholar] [CrossRef]
- Bhor, S.A.; Tateda, C.; Mochizuki, T.; Sekine, K.T.; Yaeno, T.; Yamaoka, N.; Nishiguchi, M.; Kobayashi, K. Inducible transgenic tobacco system to study the mechanisms underlying chlorosis mediated by the silencing of chloroplast heat shock protein 90. Virus Dis. 2017, 28, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Liberek, K.; Georgopoulos, C.; Zylicz, M. Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl. Acad. Sci. USA 1988, 85, 6632–6636. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bu, C.; Li, T.; Wang, S.; Jiang, F.; Yi, Y.; Yang, H.; Zhang, Z. Cloning and analysis of DnaJ family members in the silkworm, Bombyx mori. Gene 2016, 576, 88–98. [Google Scholar] [CrossRef]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Rajan, V.B.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef]
- Fan, F.; Yang, X.; Cheng, Y.; Kang, Y.; Chai, X. The DnaJ Gene Family in Pepper (Capsicum annuum L.): Comprehensive Identification, Characterization and Expression Profiles. Front. Plant Sci. 2017, 8, 689. [Google Scholar]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; dePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilgin, D.D.; Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. P58 (IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inbibitor, functions in viral pathogenesis. Dev. Cell 2003, 4, 651–661. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ren, S.; Li, Q.; Royster, A.D.; Lin, L.; Liu, S.; Ganaie, S.S.; Qiu, J.; Mir, S.; Mir, M.A. Hantaviruses use the endogenous host factor P58IPK to combat the PKR antiviral response. PLoS Pathog. 2021, 17, e1010007. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Veiga, R.; Ghita, M.; Tsikou, D.; Medina, V.; Canto, T.; Makris, A.M.; Livieratos, I.C. Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 2012, 163, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Hao, K.; Yu, F.; Shen, L.; Wang, F.; Yang, J.; Su, C. Field application of nanoliposomes delivered quercetin by inhibiting specific hsp70 gene expression against plant virus disease. J. Nanobiotechnol. 2022, 20, 16. [Google Scholar] [CrossRef]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef] [Green Version]
- Schroda, M.; Mühlhaus, T. A “foldosome” in the chloroplast? Plant Signal. Behav. 2009, 4, 301–303. [Google Scholar] [CrossRef]
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Fan, P.; Jiang, P.; Lv, S.; Chen, X.; Li, Y. Chloroplast-targeted Hsp90 plays essential roles in plastid development and embryogenesis in Arabidopsis possibly linking with VIPP1. Physiol. Plant. 2014, 150, 292–307. [Google Scholar] [CrossRef]
- Islam, S.; Bhor, S.A.; Tanaka, K.; Sakamoto, H.; Yaeno, T.; Kaya, H.; Kobayashi, K. Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-like Phenotype. Int. J. Mol. Sci. 2020, 21, 4202. [Google Scholar] [CrossRef]
- Navarro, B.; Gisel, A.; Rodio, M.E.; Delgado, S.; Flores, R.; Serio, F.D. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012, 70, 991–1003. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Chen, R.; Tu, Z.; Nie, X.; Song, B.; He, C.; Xie, C.; Nie, B. Global Screening and Functional Identification of Major HSPs Involved in PVY Infection in Potato. Genes 2022, 13, 566. https://doi.org/10.3390/genes13040566
Li K, Chen R, Tu Z, Nie X, Song B, He C, Xie C, Nie B. Global Screening and Functional Identification of Major HSPs Involved in PVY Infection in Potato. Genes. 2022; 13(4):566. https://doi.org/10.3390/genes13040566
Chicago/Turabian StyleLi, Kun, Ruhao Chen, Zheng Tu, Xianzhou Nie, Botao Song, Changzheng He, Conghua Xie, and Bihua Nie. 2022. "Global Screening and Functional Identification of Major HSPs Involved in PVY Infection in Potato" Genes 13, no. 4: 566. https://doi.org/10.3390/genes13040566
APA StyleLi, K., Chen, R., Tu, Z., Nie, X., Song, B., He, C., Xie, C., & Nie, B. (2022). Global Screening and Functional Identification of Major HSPs Involved in PVY Infection in Potato. Genes, 13(4), 566. https://doi.org/10.3390/genes13040566






