Distinct Minor Splicing Patterns across Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
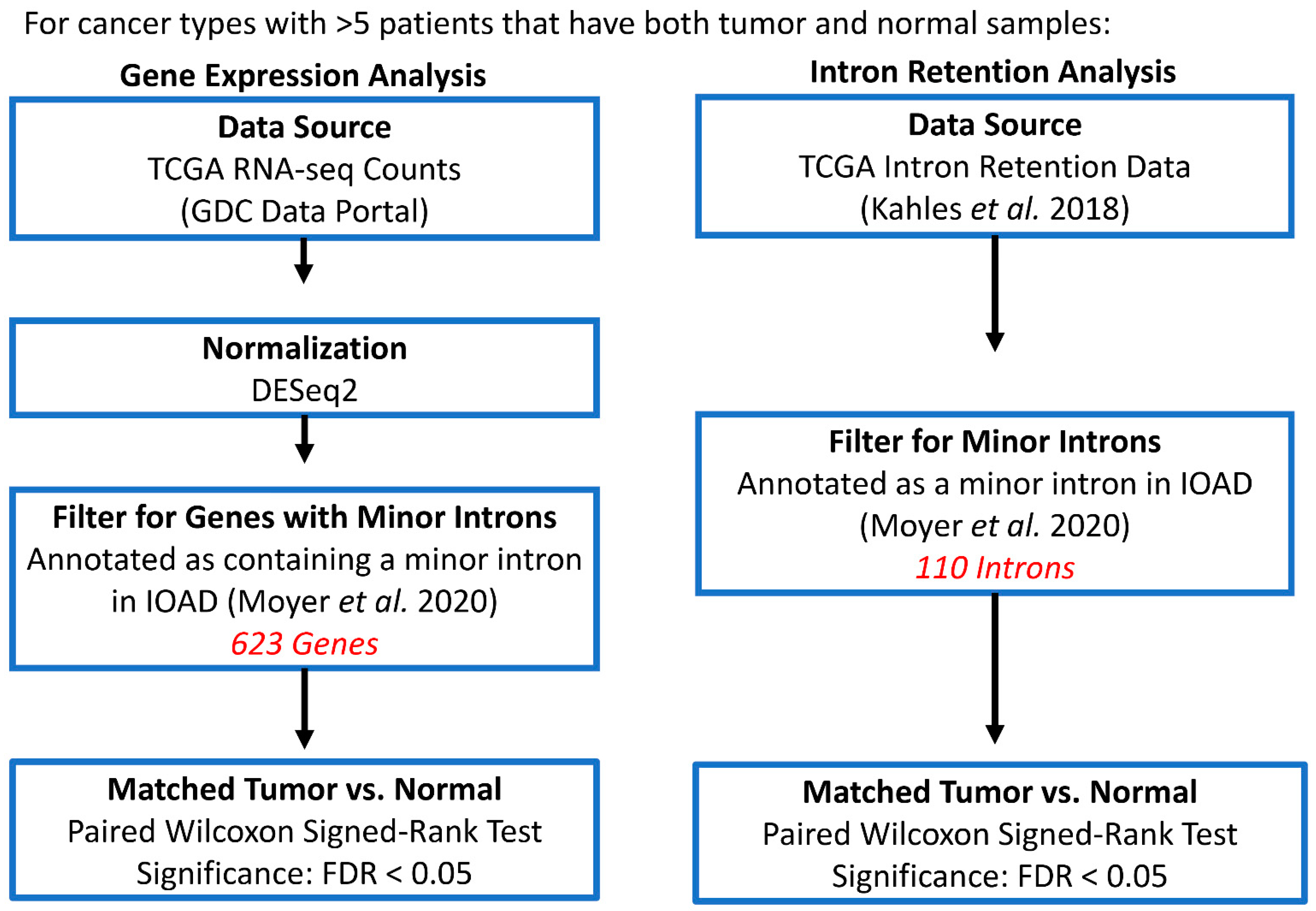
2.2. Minor Intron Retention Analysis across TCGA Cohorts
2.3. Gene Expression Analysis across TCGA Cohorts
2.4. Mutation of MIGs across TCGA Cohorts
2.5. Estimation of Minor Splicing Activity in Tumors
2.6. Estimation of Changes in Minor Splicing Activity between Matched Tumor and Normal Tissues
2.7. Functional Enrichment Analysis
2.8. Correlation of Changes in Gene Expression and Minor Splicing across TCGA Cohorts
3. Results
3.1. Minor Intron-Containing Genes Tend to Be Upregulated in Tumors
3.2. TCGA Cohorts Show Diverse Patterns of Minor Splicing
3.3. Mutations in Minor Intron-Containing Genes Do Not Correlate with Change in Minor Splicing
3.4. No Clear Patterns of Minor Splicesome Expression
3.5. Patterns in TCGA Cohorts Are Not Associated with a Particular Regulatory Netowrk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coltri, P.P.; dos Santos, M.G.P.; da Silva, G.H.G. Splicing and Cancer: Challenges and Opportunities. WIREs RNA 2019, 10, e1527. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Moore, M.J. The Spliceosome: Disorder and Dynamics Defined. Curr. Opin. Struct. Biol. 2014, 24, 141–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative Splicing as a Regulator of Development and Tissue Identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA MiR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-MRNA Splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez, D.; Tsai, Y.-H.; Weatheritt, R.; Wang, Y.; Blencowe, B.J.; Wang, Z. An Extensive Program of Periodic Alternative Splicing Linked to Cell Cycle Progression. eLife 2016, 5, e10288. [Google Scholar] [CrossRef]
- Kim, E.; Goren, A.; Ast, G. Insights into the Connection between Cancer and Alternative Splicing. Trends Genet. 2008, 24, 7–10. [Google Scholar] [CrossRef]
- Rahman, M.A.; Nasrin, F.; Bhattacharjee, S.; Nandi, S. Hallmarks of Splicing Defects in Cancer: Clinical Applications in the Era of Personalized Medicine. Cancers 2020, 12, 1381. [Google Scholar] [CrossRef]
- Oltean, S.; Bates, D.O. Hallmarks of Alternative Splicing in Cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, I.M.; Cheng, A.W.; Flytzanis, N.C.; Balsamo, M.; Condeelis, J.S.; Oktay, M.H.; Burge, C.B.; Gertler, F.B. An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype. PLoS Genet. 2011, 7, e1002218. [Google Scholar] [CrossRef] [Green Version]
- Turunen, J.J.; Niemelä, E.H.; Verma, B.; Frilander, M.J. The Significant Other: Splicing by the Minor Spliceosome. WIREs RNA 2013, 4, 61–76. [Google Scholar] [CrossRef] [Green Version]
- El Marabti, E.; Malek, J.; Younis, I. Minor Intron Splicing from Basic Science to Disease. Int. J. Mol. Sci. 2021, 22, 6062. [Google Scholar] [CrossRef] [PubMed]
- Meinke, S.; Goldammer, G.; Weber, A.I.; Tarabykin, V.; Neumann, A.; Preussner, M.; Heyd, F. Srsf10 and the Minor Spliceosome Control Tissue-Specific and Dynamic SR Protein Expression. eLife 2020, 9, e56075. [Google Scholar] [CrossRef] [PubMed]
- Turunen, J.J.; Will, C.L.; Grote, M.; Lührmann, R.; Frilander, M.J. The U11-48K Protein Contacts the 5′ Splice Site of U12-Type Introns and the U11-59K Protein. Mol. Cell Biol. 2008, 28, 3548–3560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvinge, H.; Kim, E.; Abdel-Wahab, O.; Bradley, R.K. RNA Splicing Factors as Oncoproteins and Tumor Suppressors. Nat. Rev. Cancer 2016, 16, 413–430. [Google Scholar] [CrossRef]
- Yoshida, K.; Ogawa, S. Splicing Factor Mutations and Cancer: Splicing Factor Mutations and Cancer. WIREs RNA 2014, 5, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Inoue, D.; Polaski, J.T.; Taylor, J.; Castel, P.; Chen, S.; Kobayashi, S.; Hogg, S.J.; Hayashi, Y.; Pineda, J.M.B.; El Marabti, E.; et al. Minor Intron Retention Drives Clonal Hematopoietic Disorders and Diverse Cancer Predisposition. Nat. Genet. 2021, 53, 707–718. [Google Scholar] [CrossRef]
- de Wolf, B.; Oghabian, A.; Akinyi, M.V.; Hanks, S.; Tromer, E.C.; van Hooff, J.J.E.; van Voorthuijsen, L.; van Rooijen, L.E.; Verbeeren, J.; Uijttewaal, E.C.H.; et al. Chromosomal Instability by Mutations in the Novel Minor Spliceosome Component CENATAC. EMBO J. 2021, 40, e106536. [Google Scholar] [CrossRef]
- Younis, I.; Dittmar, K.; Wang, W.; Foley, S.W.; Berg, M.G.; Hu, K.Y.; Wei, Z.; Wan, L.; Dreyfuss, G. Minor Introns Are Embedded Molecular Switches Regulated by Highly Unstable U6atac SnRNA. eLife 2013, 2, e00780. [Google Scholar] [CrossRef]
- Kahles, A.; Lehmann, K.-V.; Toussaint, N.C.; Hüser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Caesar-Johnson, S.J.; et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8705 Patients. Cancer Cell 2018, 34, 211–224.e6. [Google Scholar] [CrossRef] [Green Version]
- David, J.K.; Maden, S.K.; Weeder, B.R.; Thompson, R.F.; Nellore, A. Putatively Cancer-Specific Exon–Exon Junctions Are Shared across Patients and Present in Developmental and Other Non-Cancer Cells. NAR Cancer 2020, 2, zcaa001. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, J.; Sun, Y.; Perea-Chamblee, T.E.; Manley, J.L.; Rabadan, R. Pan-Cancer Analysis Identifies Mutations in SUGP1 That Recapitulate Mutant SF3B1 Splicing Dysregulation. Proc. Natl. Acad. Sci. USA 2020, 117, 10305–10312. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Deng, Y.; Wang, K.; Zhou, H.; Zheng, X.; Si, L.; Fu, Z. Profiles of Alternative Splicing in Colorectal Cancer and Their Clinical Significance: A Study Based on Large-Scale Sequencing Data. EBioMedicine 2018, 36, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Yang, G.; Wang, K.; Riaz, M.; Xu, J.; Lv, Z.; Zhou, H.; Li, Q.; Li, W.; Sun, J.; et al. Genome-Wide Transcriptional Analysis Reveals Alternative Splicing Event Profiles in Hepatocellular Carcinoma and Their Prognostic Significance. Front. Genet. 2020, 11, 879. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyer, D.C.; Larue, G.E.; Hershberger, C.E.; Roy, S.W.; Padgett, R.A. Comprehensive Database and Evolutionary Dynamics of U12-Type Introns. Nucleic Acids Res. 2020, 48, 7066–7078. [Google Scholar] [CrossRef] [PubMed]
- Kahles, A.; Ong, C.S.; Zhong, Y.; Rätsch, G. SplAdder: Identification, Quantification and Testing of Alternative Splicing Events from RNA-Seq Data. Bioinformatics 2016, 32, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.A.; Balanis, N.G.; Nanjundiah, A.; Sheu, K.M.; Tsai, B.L.; Zhang, Q.; Park, J.W.; Thompson, M.; Huang, J.; Witte, O.N.; et al. A Human Adult Stem Cell Signature Marks Aggressive Variants across Epithelial Cancers. Cell Rep. 2018, 24, 3353–3366.e5. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER Version 16: A Revised Family Classification, Tree-Based Classification Tool, Enhancer Regions and Extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Dvinge, H.; Bradley, R.K. Widespread Intron Retention Diversifies Most Cancer Transcriptomes. Genome Med. 2015, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Verbeeren, J.; Niemelä, E.H.; Turunen, J.J.; Will, C.L.; Ravantti, J.J.; Lührmann, R.; Frilander, M.J. An Ancient Mechanism for Splicing Control: U11 SnRNP as an Activator of Alternative Splicing. Mol. Cell 2010, 37, 821–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levesque, L.; Salazar, N.; Roy, S.W. Distinct Minor Splicing Patterns across Cancers. Genes 2022, 13, 387. https://doi.org/10.3390/genes13020387
Levesque L, Salazar N, Roy SW. Distinct Minor Splicing Patterns across Cancers. Genes. 2022; 13(2):387. https://doi.org/10.3390/genes13020387
Chicago/Turabian StyleLevesque, Lauren, Nicole Salazar, and Scott William Roy. 2022. "Distinct Minor Splicing Patterns across Cancers" Genes 13, no. 2: 387. https://doi.org/10.3390/genes13020387
APA StyleLevesque, L., Salazar, N., & Roy, S. W. (2022). Distinct Minor Splicing Patterns across Cancers. Genes, 13(2), 387. https://doi.org/10.3390/genes13020387





