An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry and Handling
2.2. Cloning of Amex Tert Gene Fragments
2.3. Completion and Sequence Analysis of the Amex TERT Amino Acid Sequence
2.4. Analysis of Amex Tert mRNA Expression
2.5. Analysis of Telomerase Activity
3. Results
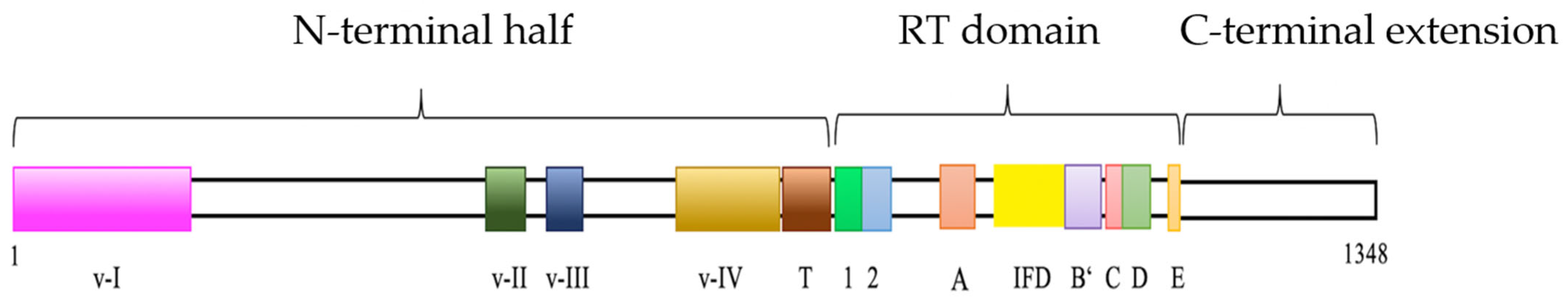
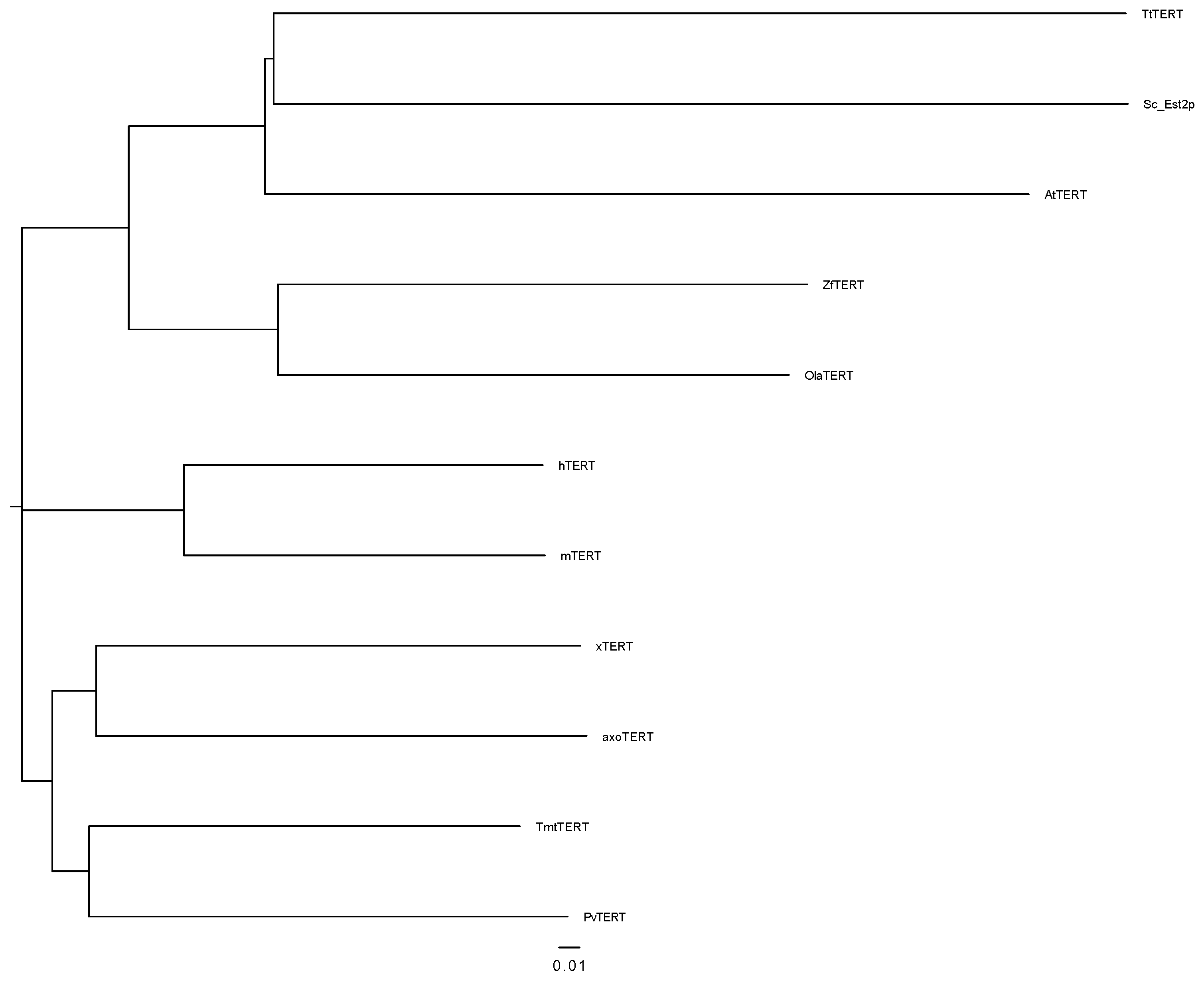
3.1. Identification and Characterization of Amex TERT
3.2. Expression Analysis of Amex Tert mRNA by RT-PCR in Somatic and Blastema Tissues
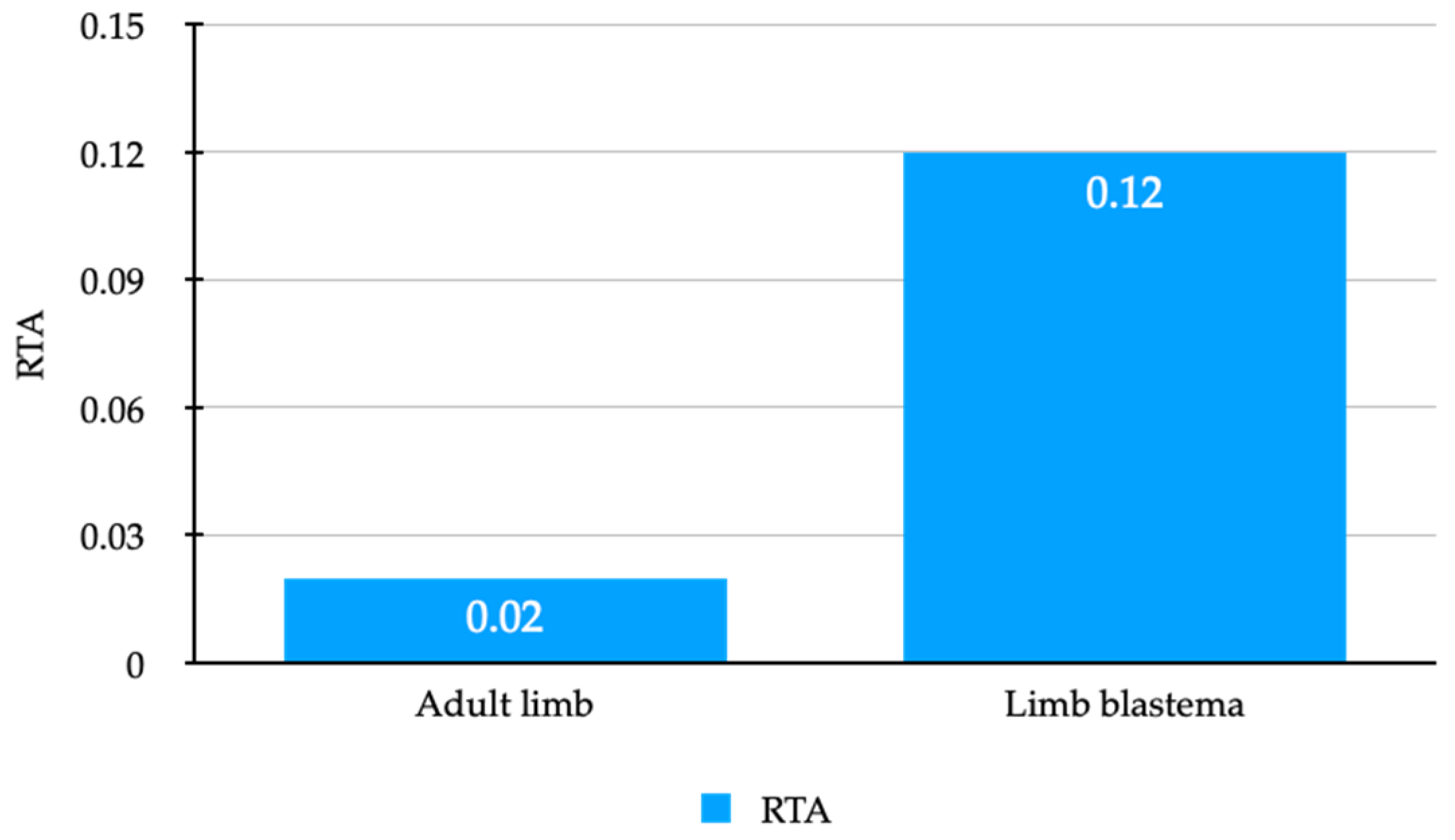
3.3. Active Telomerase Is Detectable in Axolotl Adult Limbs and Limb Blastemas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCusker, C.; Gardiner, D.M. The axolotl model for regeneration and aging research: A mini-review. Gerontology 2011, 57, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Robert Koch Institut. Demografischer Wandel. Available online: https://www.rki.de/DE/Content/Gesundheitsmonitoring/Themen/Demografischer_Wandel/Demografischer_Wandel_node.html;jsessionid=07B58C657E419E282F5153EF8AC7B88D.1_cid363 (accessed on 15 June 2019).
- Stocum, D.L. Regenerative Biology and Medicine, 2nd ed.; Regeneration of Musculoskeletal Tissues; Academic Press: Cambridge, MA, USA, 2012; Chapter 6; pp. 127–160. [Google Scholar]
- Einhorn, T.A. The cell and molecular biology of fracture healing. Clin. Orthop. Relat. Res. 1998, 355, S7–S21. [Google Scholar] [CrossRef] [PubMed]
- Allbrook, D. Skeletal muscle regeneration. Muscle Nerve 1981, 4, 234–245. [Google Scholar] [CrossRef]
- Gurtner, G.C.; Werner, S.; Barrandon, Y.; Longaker, M.T. Wound repair and regeneration. Nature 2008, 453, 314–321. [Google Scholar] [CrossRef]
- Stocum, D.L. Regenerative Biology and Medicine, 2nd ed.; An Overview of Regenerative Biology; Academic Press: Cambridge, MA, USA, 2012; Chapter 1; pp. 3–18. [Google Scholar]
- Kumar, A.; Brockes, J.P. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat. Rev. Mol. Cell Biol. 2002, 3, 566–574. [Google Scholar] [CrossRef]
- Reiß, C.; Olsson, L.; Hoßfeld, U. The history of the oldest self-sustaining laboratory animal: 150 years of axolotl research. J. Exp. Zool. B. Mol. Dev. Evol. 2015, 324, 393–404. [Google Scholar] [CrossRef]
- Joven, A.; Elewa, A.; Simon, A. Model systems for regeneration: Salamanders. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Stocum, D.L. Regenerative Biology and Medicine, 2nd ed.; Regeneration of Appendages; Academic Press: Cambridge, MA, USA, 2012; Chapter 8; pp. 183–226. [Google Scholar]
- Campbell, L.J.; Crews, C.M. Wound epidermis formation and function in urodele amphibian limb regeneration. Cell. Mol. Life Sci. 2008, 65, 73–79. [Google Scholar] [CrossRef]
- Muneoka, K.; Fox, W.F.; Bryant, S.V. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 1986, 116, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Stocum, D.L. The relation of mitotic index, cell density, and growth to pattern regulation in regenerating Ambystoma maculatum forelimbs. J. Exp. Zool. 1980, 212, 233–242. [Google Scholar] [CrossRef]
- Haas, B.J.; Whited, J.L. Advances in decoding axolotl limb regeneration. Trends Genet. 2017, 33, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Tank, P.W.; Carlson, B.M. A staging system for forelimb regeneration in the axolotl, Ambystoma mexicanum. J. Morph. 1976, 150, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.M.V.; Shay, J.W.; Wright, W.E. Telomere biology in metazoa. FEBS Lett. 2010, 584, 3741–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [Green Version]
- Harley, C.B.; Greider, C.W.; Futcher, A.B. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Sykorova, E.; Fajkus, J. Structure-function relationships in telomerase genes. Biol. Cell. 2009, 101, 375–392. [Google Scholar] [CrossRef]
- Wright, W.E.; Piatyszek, M.A.; Rainey, W.E.; Byrd, W.; Shay, J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996, 18, 173–179. [Google Scholar] [CrossRef]
- Chiu, C.P.; Dragowska, W.; Kim, N.W.; Vaziri, H.; Yui, J.; Thomas, T.E.; Harley, C.B.; Lansdorp, P.M. Differential expression of telomerase activity in hematopoietic progenitors from adult human bone marrow. Stem. Cells 1996, 14, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Martens, U.M.; Brass, V.; Sedlacek, L.; Pantic, M.; Exner, C.; Guo, Y.; Engelhardt, M.; Lansdorp, P.M.; Waller, C.F.; Lange, W. Telomere maintenance in human B lymphocytes. Br. J. Haematol. 2002, 119, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Djojosubroto, M.W.; Choi, Y.S.; Lee, H.; Rudolph, K.L. Telomeres and telomerase in aging, regeneration and cancer. Mol. Cells 2003, 15, 164–175. [Google Scholar]
- Francis, N.; Gregg, T.; Owen, R.; Ebert, T.; Bodnar, A. Lack of age-associated telomere shortening in long- and short-lived species of sea urchins. FEBS Lett. 2006, 580, 4713–4717. [Google Scholar] [CrossRef] [Green Version]
- Tan, T.C.J.; Rahman, R.; Jaber-Hijazi, F.; Felix, D.A.; Chen, C.; Louis, E.J.; Aboobaker, A. Telomere maintenance and telomerase activity are differentially regulated in asexual and sexual worms. Proc. Natl. Acad. Sci. USA 2012, 109, 4209–4214. [Google Scholar] [CrossRef] [Green Version]
- Yun, M.H. Changes in regenerative capacity through lifespan. Int. J. Mol. Sci. 2015, 16, 25392–25432. [Google Scholar] [CrossRef] [Green Version]
- Sal-Site. Ambystoma.uky.edu. Assembly V4.0. Available online: https://ambystoma.uky.edu/quick-links/est-database (accessed on 12 December 2019).
- Vieira, J.; Messing, J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987, 153, 3–11. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. National Center for Biotechnology Information, U.S. National Library of Medicine. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 12 December 2019).
- ExPASy Bioformatics Resources Portal. Swiss Institute of Bioinformatics. Available online: https://web.expasy.org/translate/ (accessed on 3 November 2018).
- Clustal Omega: Multiple Sequence Alignment. European Molecular Biology Laboratory (EMBL-EBI). Available online: https://www.ebi.ac.uk/Tools/services/web_clustalo/toolform.ebi (accessed on 23 March 2019).
- Guelke, E.; Bucan, V.; Liebsch, C.; Lazaridis, A.; Radtke, C.; Vogt, P.M.; Reimers, K. Identification of reference genes and validation for gene expression studies in diverse axolotl (ambystoma mexicanum) tissues. Gene 2015, 560, 114–123. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Herbert, B.S.; Hochreiter, A.E.; Wright, W.E.; Shay, J.W. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat. Protoc. 2006, 1, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Aldous, W.K.; Marean, A.J.; DeHart, M.J.; Matej, L.A.; Moore, K.H. Effects of tamoxifen on telomerase activity in breast carcinoma cell lines. Cancer 1999, 85, 1523–1529. [Google Scholar] [CrossRef]
- Haas, B.J.; Bryant, D.; Di Tommaso, T.; Lee, T.; Tickle, T.; Couger, M.B.; Guzikowski, A.; Tsai, S.; Coyne, S.; Freeman, R.; et al. TSA: Ambystoma Mexicanum Ambymex_c1080993_g3_i1 Transcribed RNA Sequence. National Center for Biotechnology Information, U.S. National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/nuccore/GFBM010789021.1/ (accessed on 16 November 2016).
- Springhetti, S.; Strauss, S.; Reimers, K.; Lazaridis, A.; Liebsch, C.; Vogt, P.M. Ambystoma Mexicanum Telomerase Reverse Transcriptase (TERT) mRNA, Complete cds. National Center for Biotechnology Information. U.S. National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MK702005.1 (accessed on 19 November 2019).
- Springhetti, S.; Strauss, S.; Reimers, K.; Lazaridis, A.; Liebsch, C.; Vogt, P.M. Telomerase Reverse Transcriptase [Ambystoma Mexicanum]. National Center for Biotechnology Information. U.S. National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/protein/1775571859 (accessed on 19 November 2019).
- Nakamura, T.M.; Morin, G.B.; Chapman, K.B.; Weinrich, S.L.; Andrews, W.H.; Lingner, J.; Harley, C.B.; Cech, T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Lue, N.F.; Lin, Y.; Mian, I.S. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 2003, 23, 8440–8449. [Google Scholar] [CrossRef] [Green Version]
- Hossain, S.; Singh, S.; Lue, N.F. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative "thumb" domain. J. Biol. Chem. 2002, 277, 36174–36180. [Google Scholar] [CrossRef] [Green Version]
- Kuramoto, M.; Ohsumi, K.; Kishimoto, T.; Ishikawa, F. Identification and analyses of the xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene 2001, 277, 101–110. [Google Scholar] [CrossRef]
- Chen, C.; Sung, T.; Chen, L.; Chen, J. Telomere maintenance during anterior regeneration and aging in the freshwater annelid aeolosoma viride. Sci. Rep. 2018, 8, 18078. [Google Scholar] [CrossRef] [Green Version]
- Lau, B.W.; Wong, A.O.; Tsao, G.S.; So, K.; Yip, H.K. Molecular cloning and characterization of the zebrafish (danio rerio) telomerase catalytic subunit (telomerase reverse transcriptase, TERT). J. Mol. Neurosci. 2008, 34, 63–75. [Google Scholar] [CrossRef]
- Pfennig, F.; Kind, B.; Zieschang, F.; Busch, M.; Gutzeit, H.O. Tert expression and telomerase activity in gonads and somatic cells of the japanese medaka (oryzias latipes). Dev. Growth Differ. 2008, 50, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Alibardi, L. Immunodetection of telomerase-like immunoreactivity in normal and regenerating tail of amphibians suggests it is related to their regenerative capacity. J. Exp. Zool. A Ecol. Genet. Physiol. 2015, 323, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Anchelin, M.; Murcia, L.; Alcaraz-Pérez, F.; García-Navarro, E.M.; Cayuela, M.L. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS ONE 2011, 6, e16955. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Yakushiji, N.; Nakada, Y.; Satoh, A.; Ide, H.; Tamura, K. Limb regeneration in xenopus laevis froglet. Sci. World J. 2006, 6 (Suppl. 1), 26–37. [Google Scholar] [CrossRef] [Green Version]
- Galliot, B.; Ghila, L. Cell plasticity in homeostasis and regeneration. Mol. Reprod. Dev. 2010, 77, 837–855. [Google Scholar] [CrossRef] [Green Version]
- Blasco, M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007, 3, 640–649. [Google Scholar] [CrossRef]
- Muneoka, K.; Allan, C.H.; Yang, X.; Lee, J.; Han, M. Mammalian regeneration and regenerative medicine. Birth Defects Res. C Embryo Today 2008, 84, 265–280. [Google Scholar] [CrossRef]
- Seifert, A.W.; Muneoka, K. The blastema and epimorphic regeneration in mammals. Dev. Biol. 2018, 433, 190–199. [Google Scholar] [CrossRef]
- Samper, E.; Flores, J.M.; Blasco, M.A. Restoration of telomerase activity rescues chromosomal instability and premature aging in terc −/− mice with short telomeres. EMBO Rep. 2001, 2, 800–807. [Google Scholar] [CrossRef] [Green Version]
| Amex TERT Average Ct | Ornithine Average Ct | △Ct (Ct Amex TERT − Ct Ornithin) | △△Ct (△Ct Blastema − △Ct Limb) | Fold Difference (2−ΔΔCt) | |
|---|---|---|---|---|---|
| Adult limb (n = 7) | 31.19 ± 1.81 | 31 ± 1.67 | 0.19 ± 2.46 | 0 | 1 |
| Limb blastema (n = 6) | 30.42 ± 1.22 | 30.93 ± 1.53 | −0.51 ± 1.96 | −0.7 ± 1.96 | 1.62 (0.42–6.32) |
| Average Ct | RTA | |
|---|---|---|
| Adult limb (n = 4) | 26.6 ± 2.23 | 0.02 (0.01–0.05) |
| Limb blastema (n = 3) | 22.02 ± 1.69 | 0.12 (0.06–0.23) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Springhetti, S.; Bucan, V.; Liebsch, C.; Lazaridis, A.; Vogt, P.M.; Strauß, S. An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT). Genes 2022, 13, 373. https://doi.org/10.3390/genes13020373
Springhetti S, Bucan V, Liebsch C, Lazaridis A, Vogt PM, Strauß S. An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT). Genes. 2022; 13(2):373. https://doi.org/10.3390/genes13020373
Chicago/Turabian StyleSpringhetti, Sina, Vesna Bucan, Christina Liebsch, Andrea Lazaridis, Peter Maria Vogt, and Sarah Strauß. 2022. "An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT)" Genes 13, no. 2: 373. https://doi.org/10.3390/genes13020373
APA StyleSpringhetti, S., Bucan, V., Liebsch, C., Lazaridis, A., Vogt, P. M., & Strauß, S. (2022). An Identification and Characterization of the Axolotl (Ambystoma mexicanum, Amex) Telomerase Reverse Transcriptase (Amex TERT). Genes, 13(2), 373. https://doi.org/10.3390/genes13020373







