A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken
Abstract
:1. Introduction
2. Materials and Methods
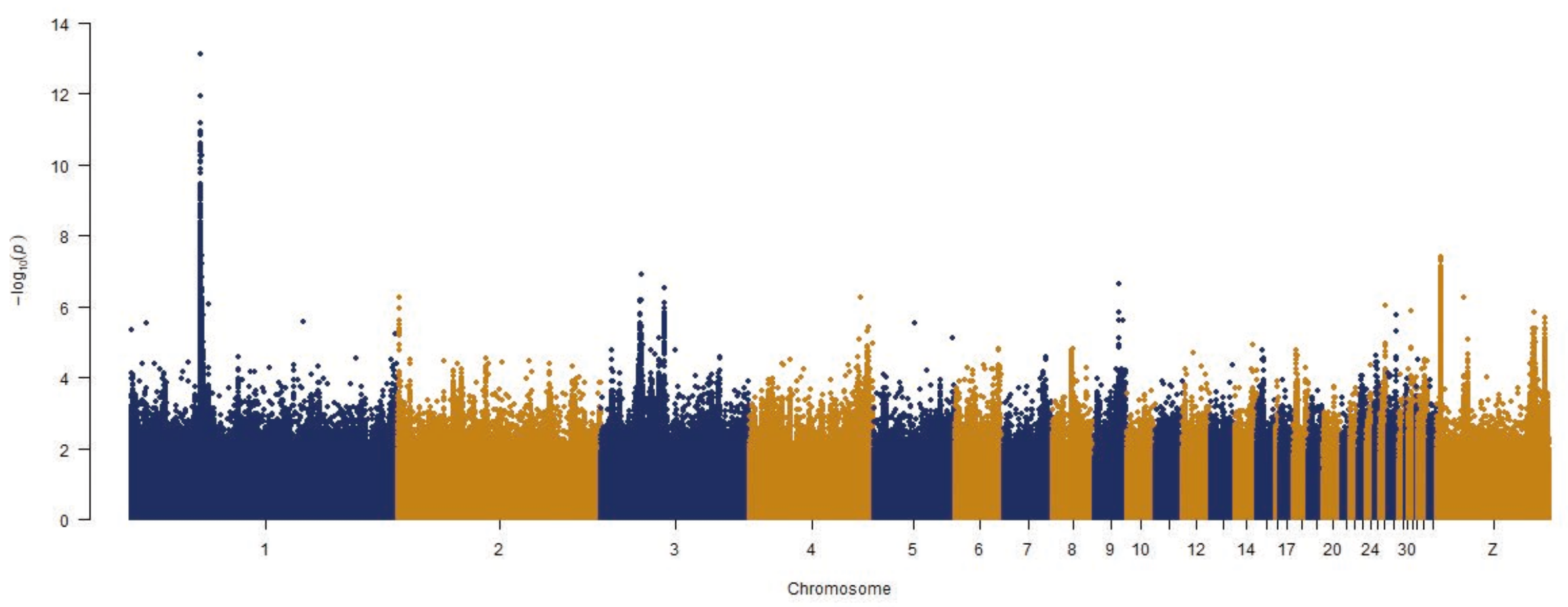
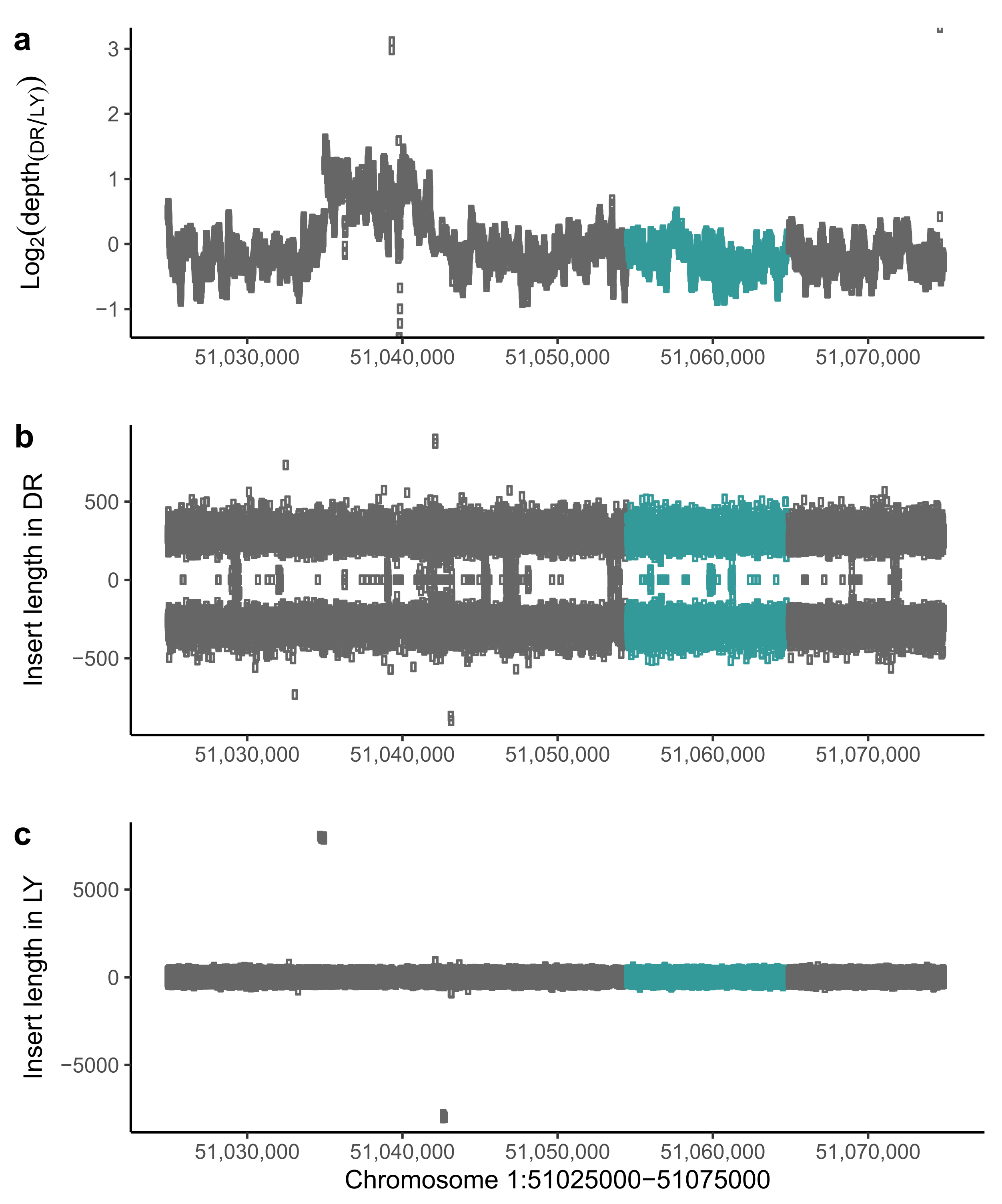
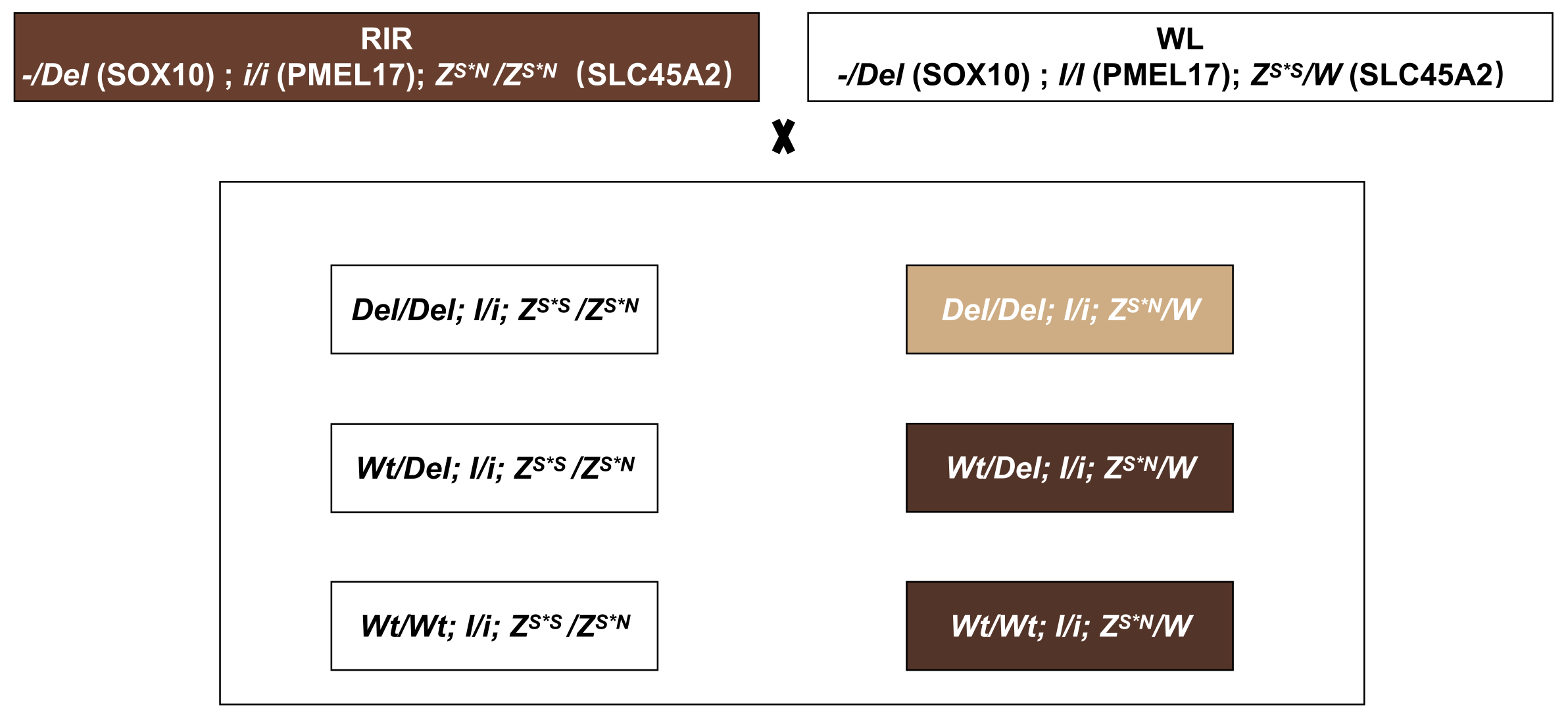
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DR | dark red |
| LY | light yellow |
| GWAS | genome wide association study |
| RIR | Rhode Island Red |
| WL | White Leghorn |
References
- Hillel, J.; Groenen, M.; Tixier-Boichard, M.; Korol, A.; David, L.; Kirzhner, V.; Burke, T.; Barre-Dirie, A.; Crooijmans, R.; Elo, K.; et al. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet. Sel. Evol. GSE 2003, 35, 533–557. [Google Scholar] [CrossRef] [Green Version]
- Xunhe, H.; Wu, Y.J.; Miao, Y.W.; Peng, M.S.; Chen, X.; He, D.L.; Suwannapoom, C.; Du, B.W.; Li, X.Y.; Weng, Z.X.; et al. Was chicken domesticated in northern China? New evidence from mitochondrial genomes. Sci. Bull. 2018, 63, 743–746. [Google Scholar] [CrossRef] [Green Version]
- Moiseyeva, I.G.; Romanov, M.N.; Nikiforov, A.A.; Sevastyanova, A.A.; Semyenova, S.K. Evolutionary relationships of Red Jungle Fowl and chicken breeds. Genet. Sel. Evol. 2003, 35, 403. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.S.; Thakur, M.; Peng, M.S.; Jiang, Y.; Frantz, L.A.F.; Li, M.; Zhang, J.J.; Wang, S.; Peters, J.; Otecko, N.O.; et al. 863 genomes reveal the origin and domestication of chicken. Cell Res. 2020, 30, 693–701. [Google Scholar] [CrossRef]
- Agnvall, B.; Bélteky, J.; Jensen, P. Brain size is reduced by selection for tameness in Red Junglefowl– correlated effects in vital organs. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Katajamaa, R.; Jensen, P. Tameness correlates with domestication related traits in a Red Junglefowl intercross. Genes Brain Behav. 2021, 20, e12704. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, D.A.; Honaker, C.; Dorshorst, B.; Andersson, L.; Brisbin, I.; Siegel, P. Growth patterns for three generations of an intercross between red junglefowl and chickens selected for low body weight. J. Anim. Breed. Genet. 2018, 135. [Google Scholar] [CrossRef] [PubMed]
- Saino, N.; Romano, M.; Rubolini, D.; Teplitsky, C.; Ambrosini, R.; Caprioli, M.; Canova, L.; Wakamatsu, K. Sexual Dimorphism in Melanin Pigmentation, Feather Coloration and Its Heritability in the Barn Swallow (Hirundo rustica). PLoS ONE 2013, 8, e58024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoddard, M.C.; Prum, R.O. How colorful are birds? Evolution of the avian plumage color gamut. Behav. Ecol. 2011, 22, 1042–1052. [Google Scholar] [CrossRef] [Green Version]
- Aspengren, S.; Sköld, H.N.; Quiroga, G.; Mårtensson, L.; Wallin, M. Noradrenaline- and Melatonin-Mediated Regulation of Pigment Aggregation in Fish Melanophores. Pigment. Cell Res. 2003, 16, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hearing, V.J.; Tsukamoto, K.; Urabe, K.; Kameyama, K.; Montague, P.M.; Jackson, I.j. Functional Properties of Cloned Melanogenic Proteins. Pigment. Cell Res. 1992, 5, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Fogelholm, J.; Henriksen, R.; Höglund, A.; Huq, N.; Johnsson, M.; Lenz, R.; Jensen, P.; Wright, D. CREBBP and WDR 24 Identified as Candidate Genes for Quantitative Variation in Red-Brown Plumage Colouration in the Chicken. Sci. Rep. 2020, 10, 1161. [Google Scholar] [CrossRef]
- Huang, T.; Pu, Y.; Song, C.; Sheng, Z.; Hu, X. A quantitative trait locus on chromosome 2 was identified that accounts for a substantial proportion of phenotypic variance of the yellow plumage color in chicken. Poult. Sci. 2020, 99, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bed Hom, B.; Marthey, S.; Valade, M.; Dureux, A.; Moroldo, M.; Péchoux, C.; Coville, J.L.; Gourichon, D.; Vieaud, A.; et al. A missense mutation in TYRP1 causes the chocolate plumage color in chicken and alters melanosome structure. Pigment. Cell Melanoma Res. 2019, 32, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Kerje, S.; Sharma, P.; Gunnarsson, U.; Kim, H.; Bagchi, S.; Fredriksson, R.; Schütz, K.; Jensen, P.; von Heijne, G.; Okimoto, R.; et al. The Dominant white, Dun and Smoky Color Variants in Chicken Are Associated With Insertion/Deletion Polymorphisms in the PMEL17 GeneSequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY636124, AY636125, AY636126, AY636127, AY636128, AY636129. Genetics 2004, 168, 1507–1518. [Google Scholar] [CrossRef] [Green Version]
- Gunnarsson, U.; Hellstrom, A.R.; Tixier-Boichard, M.; Minvielle, F.; Bed’hom, B.; Ito, S.; Jensen, P.; Rattink, A.; Vereijken, A.; Andersson, L. Mutations in SLC45A2 Cause Plumage Color Variation in Chicken and Japanese Quail. Genetics 2007, 175, 867–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araguas, R.M.; Sanz, N.; Viñas, J.; Vidal, O. MC1R polymorphism associated with plumage color variations in Coturnix chinensis. Anim. Genet. 2018, 49, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Li, X.L.; Li, Y.; Gu, Z.L.; Zheng, C.S.; Wei, Z.H.; Wang, J.S.; Zhou, R.Y.; Li, L.H.; Zheng, H.Q. Genetic variation of chicken MC1R gene in different plumage colour populations. Br. Poult. Sci. 2010, 51, 734–739. [Google Scholar] [CrossRef]
- Kerje, S.; Lind, J.; Schütz, K.; Jensen, P.; Andersson, L. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim. Genet. 2003, 34, 241–248. [Google Scholar] [CrossRef]
- Gunnarsson, U.; Kerje, S.; Bed’Hom, B.; Sahlqvist, A.S.; Ekwall, O.; Tixier-Boichard, M.; Kampe, O.; Andersson, L. The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment. Cell Melanoma Res. 2011, 24, 268–274. [Google Scholar] [CrossRef]
- Schwochow Thalmann, D.; Ring, H.; Sundström, E.; Cao, X.; Larsson, M.; Kerje, S.; Höglund, A.; Fogelholm, J.; Wright, D.; Jemth, P.; et al. The evolution of Sex-linked barring alleles in chickens involves both regulatory and coding changes in CDKN2A. PLoS Genet. 2017, 13, 1–22. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Fisher, D. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Domyan, E.T.; Guernsey, M.W.; Kronenberg, Z.; Krishnan, S.; Boissy, R.E.; Vickrey, A.I.; Rodgers, C.; Cassidy, P.; Leachman, S.A.; Fondon, J.W.; et al. Epistatic and Combinatorial Effects of Pigmentary Gene Mutations in the Domestic Pigeon. Curr. Biol. 2014, 24, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonellis, A.; Bennett, W.R.; Menheniott, T.R.; Prasad, A.B.; Lee-Lin, S.Q.; Green, E.D.; Paisley, D.; Kelsh, R.N.; Pavan, W.J.; Ward Andrew, N.C.S.P. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg—Shah (WS4) syndrome. Hum. Mol. Genet. 2005, 15, 259–271. [Google Scholar] [CrossRef]
- Antonellis, A.; Huynh, J.; Lee-Lin, S.Q.; Vinton, R.; Renaud, G.; Loftus, S.; Elliot, G.; Wolfsberg, T.; Green, E.; Mccallion, A.; et al. Identification of Neural Crest and Glial Enhancers at the Mouse Sox10 Locus through Transgenesis in Zebrafish. PLoS Genet. 2008, 4, e1000174. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, T.; Liu, M.; Peng, S.; Zhang, X.; Chen, Y.; Lv, X.; Yang, W.; Li, K.; Zhang, J.; Wang, H.; et al. A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken. Genes 2022, 13, 327. https://doi.org/10.3390/genes13020327
Zhu T, Liu M, Peng S, Zhang X, Chen Y, Lv X, Yang W, Li K, Zhang J, Wang H, et al. A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken. Genes. 2022; 13(2):327. https://doi.org/10.3390/genes13020327
Chicago/Turabian StyleZhu, Tao, Mengchao Liu, Shan Peng, Xinye Zhang, Yu Chen, Xueze Lv, Weifang Yang, Kaiyang Li, Jianwei Zhang, Huie Wang, and et al. 2022. "A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken" Genes 13, no. 2: 327. https://doi.org/10.3390/genes13020327
APA StyleZhu, T., Liu, M., Peng, S., Zhang, X., Chen, Y., Lv, X., Yang, W., Li, K., Zhang, J., Wang, H., Li, H., Ning, Z., Wang, L., & Qu, L. (2022). A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken. Genes, 13(2), 327. https://doi.org/10.3390/genes13020327





