The Preliminary Evaluation of Epigenetic Modifications Regulating the Expression of IL10 in Insulin-Resistant Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human and Mouse Cell Culture
2.2. Human and Mouse Cell Line Culture, Differentiation and Insulin Resistance Induction
2.3. Insulin Resistance Induction
2.4. DNA and RNA Isolation
2.5. Reverse Transcription Reaction and Gene Expression Level
2.6. Site-Specific DNA Methylation of Cells from Cell Cultures
2.7. Global and Site-Specific Histone Modifications
2.8. Statistical Analysis
3. Results
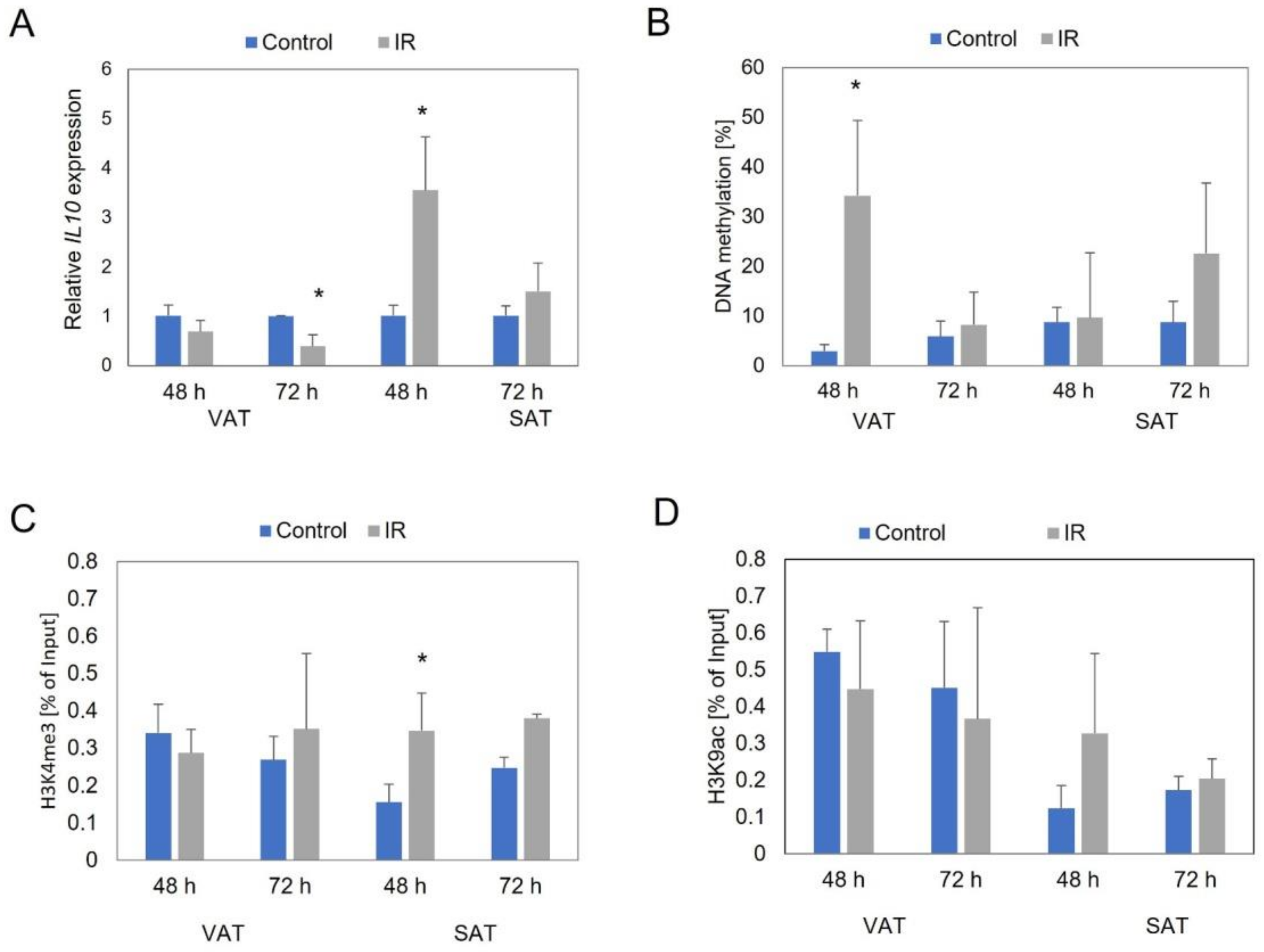
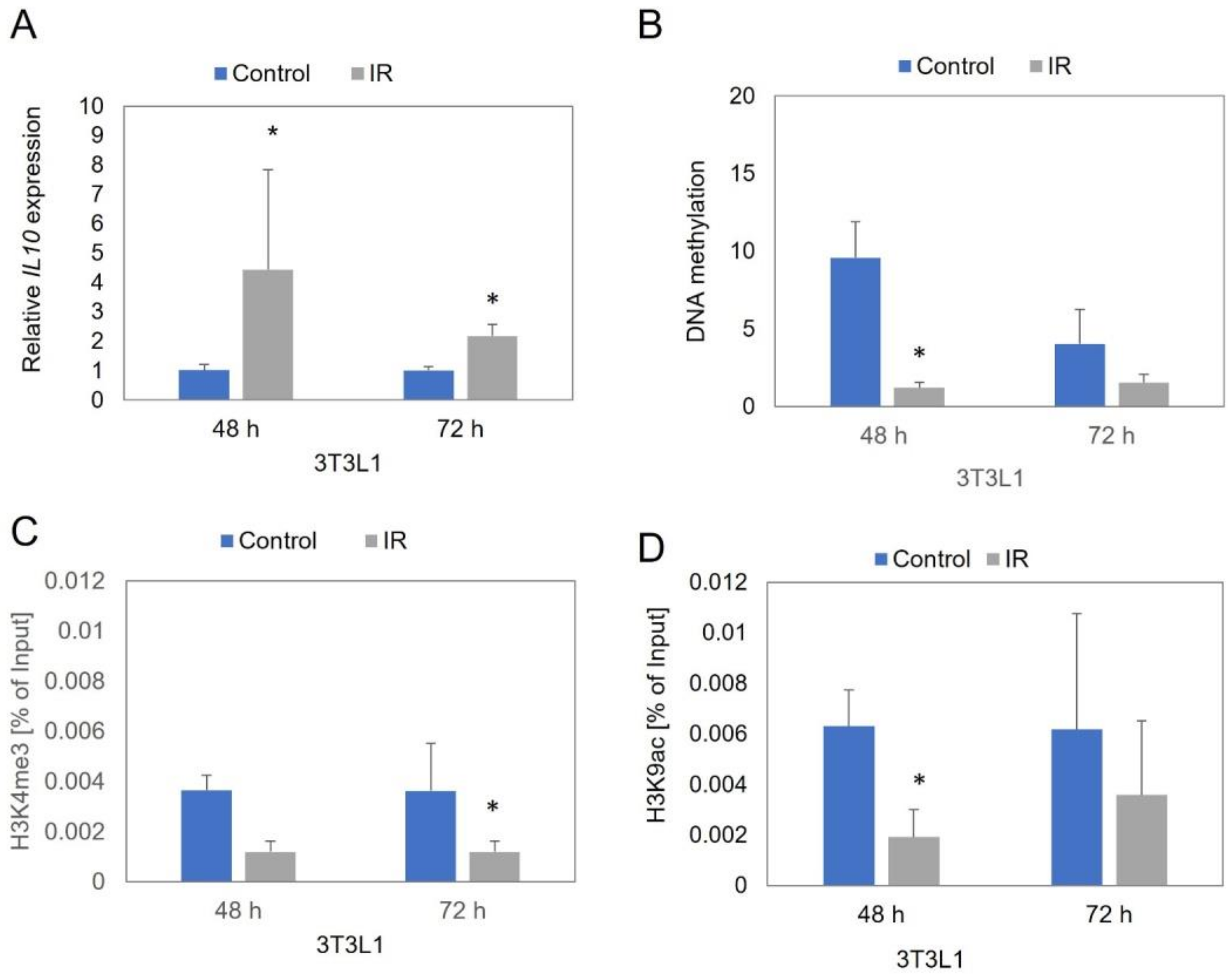
3.1. IL10 Expression Level
3.2. IL10 Promoter Region Methylation Level
3.3. Site-Specific Histone Modifications near IL10 Gene
3.4. Correlation Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aung, K.; Lorenzo, C.; Hinojosa, M.A.; Haffner, S.M. Risk of Developing Diabetes and Cardiovascular Disease in Metabolically Unhealthy Normal-Weight and Metabolically Healthy Obese Individuals. J. Clin. Endocrinol. Metab. 2014, 99, 462–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.M.; El-Serag, H.B. The Epidemiology of Obesity. Gastroenterol. Clin. N. Am. 2010, 39, 1. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hotamisligil, G.S. Obesity-Induced Inflammatory Changes in Adipose Tissue. J. Clin. Investig. 2003, 112, 1785–1788. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, L.; Roszer, T.; Ricote, M. Inflammatory Mediators and Insulin Resistance in Obesity: Role of Nuclear Receptor Signaling in Macrophages. Mediat. Inflamm. 2010, 2010, 219583. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ballantyne, C.M. Metabolic Inflammation and Insulin Resistance in Obesity. Circ. Res. 2020, 126, 1549–1564. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity is Associated with Macrophage Accumulation in Adipose Tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Verboven, K.; Wouters, K.; Gaens, K.; Hansen, D.; Bijnen, M.; Wetzels, S.; Stehouwer, C.D.; Goossens, G.H.; Schalkwijk, C.G.; Blaak, E.E.; et al. Abdominal Subcutaneous and Visceral Adipocyte Size, Lipolysis and Inflammation Relate to Insulin Resistance in Male Obese Humans. Sci. Rep. 2018, 8, 4677. [Google Scholar] [CrossRef]
- Fillatreau, S.; Gray, D.; Anderton, S.M. Not Always the Bad Guys: B Cells as Regulators of Autoimmune Pathology. Nat. Rev. Immunol. 2008, 8, 391–397. [Google Scholar] [CrossRef]
- MacKenzie, K.F.; Pattison, M.J.; Arthur, J.S.C. Transcriptional Regulation of IL-10 and Its Cell-Specific Role in Vivo. Crit. Rev. Immunol. 2014, 34, 315–345. [Google Scholar] [CrossRef]
- Juge-Aubry, C.E.; Somm, E.; Pernin, A.; Alizadeh, N.; Giusti, V.; Dayer, J.-M.; Meier, C.A. Adipose Tissue Is a Regulated Source of Interleukin-10. Cytokine 2005, 29, 270–274. [Google Scholar]
- Acosta, J.R.; Tavira, B.; Douagi, I.; Kulyté, A.; Arner, P.; Rydén, M.; Laurencikiene, J. Human-Specific Function of IL-10 in Adipose Tissue Linked to Insulin Resistance. J. Clin. Endocrinol. Metab. 2019, 104, 4552–4562. [Google Scholar] [CrossRef]
- Esposito, K.; Pontillo, A.; Giugliano, F.; Giugliano, G.; Marfella, R.; Nicoletti, G.; Giugliano, D. Association of Low Interleukin-10 Levels with the Metabolic Syndrome in Obese Women. J. Clin. Endocrinol. Metab. 2003, 88, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.G.; Hwi, J.K.; Cho, Y.R.; Kim, H.J.; Ma, Z.; Yu, T.Y.; Friedline, R.H.; Kurt-Jones, E.; Finberg, R.; Fischer, M.A.; et al. Interleukin-10 Prevents Diet-Induced Insulin Resistance by Attenuating Macrophage and Cytokine Response in Skeletal Muscle. Diabetes 2009, 58, 2525–2535. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Zhang, C.; Ma, Y.; Bu, L.; Yan, L.; Liu, D. Hydrodynamic Delivery of MIL10 Gene Protects Mice From High-Fat Diet-Induced Obesity and Glucose Intolerance. Mol. Ther. 2013, 21, 1852–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagdeviren, S.; Jung, D.Y.; Lee, E.; Friedline, R.H.; Noh, H.L.; Kim, J.H.; Patel, P.R.; Tsitsilianos, N.; Tsitsilianos, A.V.; Tran, D.A.; et al. Altered Interleukin-10 Signaling in Skeletal Muscle Regulates Obesity-Mediated Inflammation and Insulin Resistance. Mol. Cell. Biol. 2016, 36, 2956–2966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, L.; Fu, Q.; Ortega, T.M.; Zhou, L.; Rasmussen, D.; O’Keefe, J.; Zhang, K.K.; Chapes, S.K. Overexpression of IL-10 in C2D macro-phages promotes a macrophage phenotypic switch in adipose tissue environments. PLoS ONE 2014, 9, e86541. [Google Scholar]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.J.; Ryckman, K.K. Epigenetic and Developmental Influences on the Risk of Obesity, Diabetes, and Metabolic Syndrome. Diabetes Metab. Syndr. Obes. 2015, 8, 295–302. [Google Scholar]
- Małodobra-Mazur, M.; Cierzniak, A.; Pawełka, D.; Kaliszewski, K.; Rudnicki, J.; Dobosz, T. Metabolic Differences between Subcutaneous and Visceral Adipocytes Differentiated with an Excess of Saturated and Monounsaturated Fatty Acids. Genes 2020, 11, 1092. [Google Scholar] [CrossRef]
- Małodobra-Mazur, M.; Cierzniak, A.; Dobosz, T. Oleic acid influences the adipogenesis of 3T3-L1 cells via DNA Methylation and may predispose to obesity and obesity-related disorders. Lipids Health Dis. 2019, 18, 230. [Google Scholar] [CrossRef] [Green Version]
- Małodobra-Mazur, M.; Cierzniak, A.; Kaliszewski, K.; Dobosz, T. PPARG Hypermethylation as the First Epigenetic Modification in Newly Onset Insulin Resistance in Human Adipocytes. Genes 2021, 12, 889. [Google Scholar] [CrossRef]
- Epps-Fung, M.V.; Williford, J.; Wells, A.; Hardy, R.H. Fatty Acid-Induced Insulin Resistance in Adipocytes. Endocrinology 1997, 138, 4338–4345. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, Silencing Potential and Evolutionary Impact of Promoter DNA Methylation in the Human Genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.A.; Foulds, C.E.; O’Malley, B.W. Histone Marks in the ‘Drivers Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017, 42, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to de Novo Methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otani, J.; Nankumo, T.; Arita, K.; Inamoto, S.; Ariyoshi, M.; Shirakawa, M. Structural Basis for Recognition of H3K4 Methylation Status by the DNA Methyltransferase 3A ATRX–DNMT3–DNMT3L Domain. EMBO Rep. 2009, 10, 1235–1241. [Google Scholar] [CrossRef] [Green Version]
| Organism | Application | Gene | Forward Sequence | Reverse Sequence | Product Length (Base Pair) | Amount of CpG Sites * |
|---|---|---|---|---|---|---|
| Human | Gene expression | IL10 | GGACTTTAAGGGTTACCTGG | CTGGGTCTTGGTTCTCAGC | 95 | - |
| Human | Gene expression | ACTB | GAGAAGATGACCCAGATCA | TAGCACAGCCTGGATAGCAA | 72 | - |
| Human | meDIP/ChIP | IL10 | ACTGCTCTGTTGCCTGGTC | GTCTTCACTCTGCTGAAGG | 144 | 4 |
| 3T3L1 | Gene expression | IL10 | TAAGGGTTACTTGGGTTGCC | CGCATCCTGAGGGTCTTCA | 144 | - |
| 3T3L1 | Gene expression | ACTB | CCCAGATCATGTTTGAGACC | CTGGATGGCTACGTACATG | 53 | - |
| 3T3L1 | meDIP/CHIP | IL10 | CTTGCTCTTGCACTACCAAAG | TCCTCATGCCAGTCAGTAAG | 108 | 2 |
| SAT | IL10 Expression | IL10 Promoter Region Methylation | H3K4me3 | H3K9/14ac | |||||
|---|---|---|---|---|---|---|---|---|---|
| 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | ||
| IL10 expression | 48 h | - | - | - | - | r = 0.6693 p = 0.146 | r = 0.8778 p = 0.021 | - | |
| 72 h | - | - | - | - | - | r = 0.5371 p = 0.272 | - | - | |
| IL10 promoter region methylation | 48 h | - | - | - | - | r = −0.9015 p = 0.037 | - | - | - |
| 72 h | - | - | - | - | - | r = 0.6513 p = 0.161 | - | r = −0.6484 p = 0.115 | |
| H3K4me3 | 48 h | r = 0.6693 p = 0.146 | - | r = −0.9015 p = 0.037 | - | - | - | - | |
| 72 h | r = 0.5371 p = 0.272 | - | - | r = 0.6513 p = 0.161 | - | - | - | - | |
| H3K9/14ac | 48 h | r = 0.8778 p = 0.021 | - | - | - | - | - | - | - |
| 72 h | - | - | - | r = −0.6484 p = 0.115 | - | - | - | - | |
| VAT | IL10 expression | IL10 promoter region methylation | H3K4me3 | H3K9/14ac | |||||
| 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | ||
| IL10 expression | 48 h | - | - | - | - | - | - | r = 0.6812 p = 0.136 | - |
| 72 h | - | - | - | - | - | r = −0.6196 p = 0.138 | - | - | |
| IL10 promoter region methylation | 48 h | - | - | - | - | - | - | - | - |
| 72 h | - | - | - | - | - | r = −0.6372 p = 0.124 | - | r = −0.5252 p = 0.181 | |
| H3K4me3 | 48 h | - | - | - | - | - | - | r = 0.5735 p = 0.137 | - |
| 72 h | - | r = −0.6196 p = 0.138 | - | r = −0.6372 p = 0.124 | - | - | - | r = 0.4839 p = 0.271 | |
| H3K9/14ac | 48 h | r = 0.6812 p = 0.136 | - | - | - | r = 0.5735 p = 0.137 | - | - | - |
| 72 h | - | - | - | r = −0.5252 p = 0.181 | - | r = 0.4839 p = 0.271 | - | - | |
| 3T3L1 | IL10 expression | IL10 promoter region methylation | H3K4me3 | H3K9/14ac | |||||
| 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | ||
| IL10 expression | 48 h | - | - | - | - | r = −0.6842 p = 0.061 | - | - | - |
| 72 h | - | - | - | - | - | - | - | r = 0.8949 p = 0.016 | |
| IL10 promoter region methylation | 48 h | - | - | - | - | r = 0.4210 p = 0.299 | - | r = 0.8333 p = 0.010 | - |
| 72 h | - | - | - | - | - | - | - | - | |
| H3K4me3 | 48 h | r = −0.6842 p = 0.061 | - | r = 0.4210 p = 0.299 | - | - | - | - | - |
| 72 h | - | - | - | - | - | - | - | r = 0.4767 p = 0.232 | |
| H3K9/14ac | 48 h | - | - | r = 0.8333 p = 0.010 | - | - | - | - | - |
| 72 h | - | r = 0.8949 p = 0.016 | - | - | - | r = 0.4767 p = 0.232 | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cierzniak, A.; Kaliszewski, K.; Małodobra-Mazur, M. The Preliminary Evaluation of Epigenetic Modifications Regulating the Expression of IL10 in Insulin-Resistant Adipocytes. Genes 2022, 13, 294. https://doi.org/10.3390/genes13020294
Cierzniak A, Kaliszewski K, Małodobra-Mazur M. The Preliminary Evaluation of Epigenetic Modifications Regulating the Expression of IL10 in Insulin-Resistant Adipocytes. Genes. 2022; 13(2):294. https://doi.org/10.3390/genes13020294
Chicago/Turabian StyleCierzniak, Aneta, Krzysztof Kaliszewski, and Małgorzata Małodobra-Mazur. 2022. "The Preliminary Evaluation of Epigenetic Modifications Regulating the Expression of IL10 in Insulin-Resistant Adipocytes" Genes 13, no. 2: 294. https://doi.org/10.3390/genes13020294
APA StyleCierzniak, A., Kaliszewski, K., & Małodobra-Mazur, M. (2022). The Preliminary Evaluation of Epigenetic Modifications Regulating the Expression of IL10 in Insulin-Resistant Adipocytes. Genes, 13(2), 294. https://doi.org/10.3390/genes13020294







