Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Participant Characteristics
2.3. Anthropometry and Body Composition
2.4. Physical Activity
2.5. Dietary Habits
2.6. Sample Collection
2.7. Short-Chain Fatty Acids
2.8. DNA Extraction
2.9. PPARD and PPARGC1A Genotyping
2.10. Sequencing and Bioinformatics
2.11. Statistical Analysis
3. Results
3.1. Subjects, Genotypes and Allelic Frequencies
3.2. Body Composition, Physical Activity and Dietary Habits
3.3. Short-Chain Fatty Acids
3.4. Fecal Microbiota
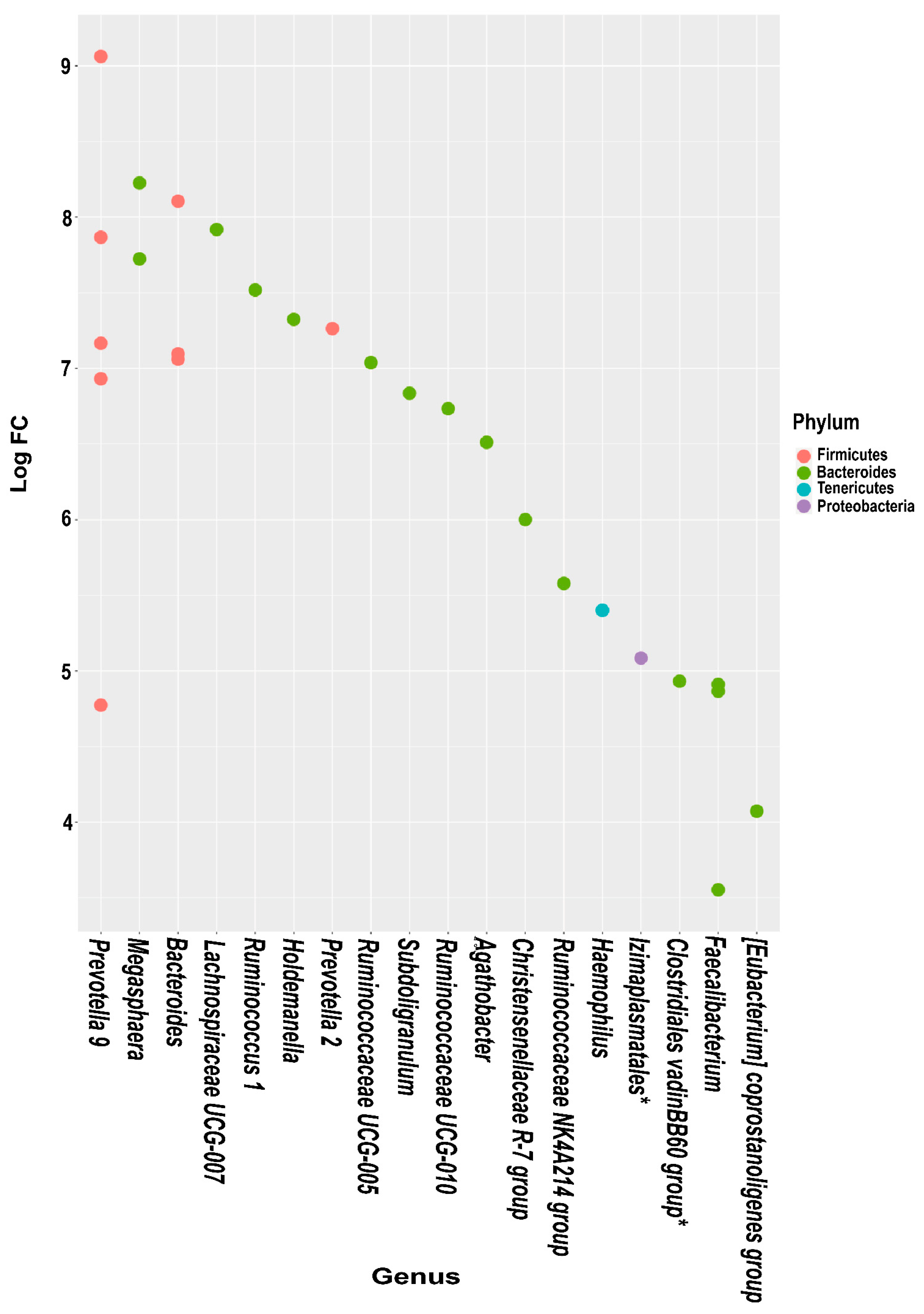
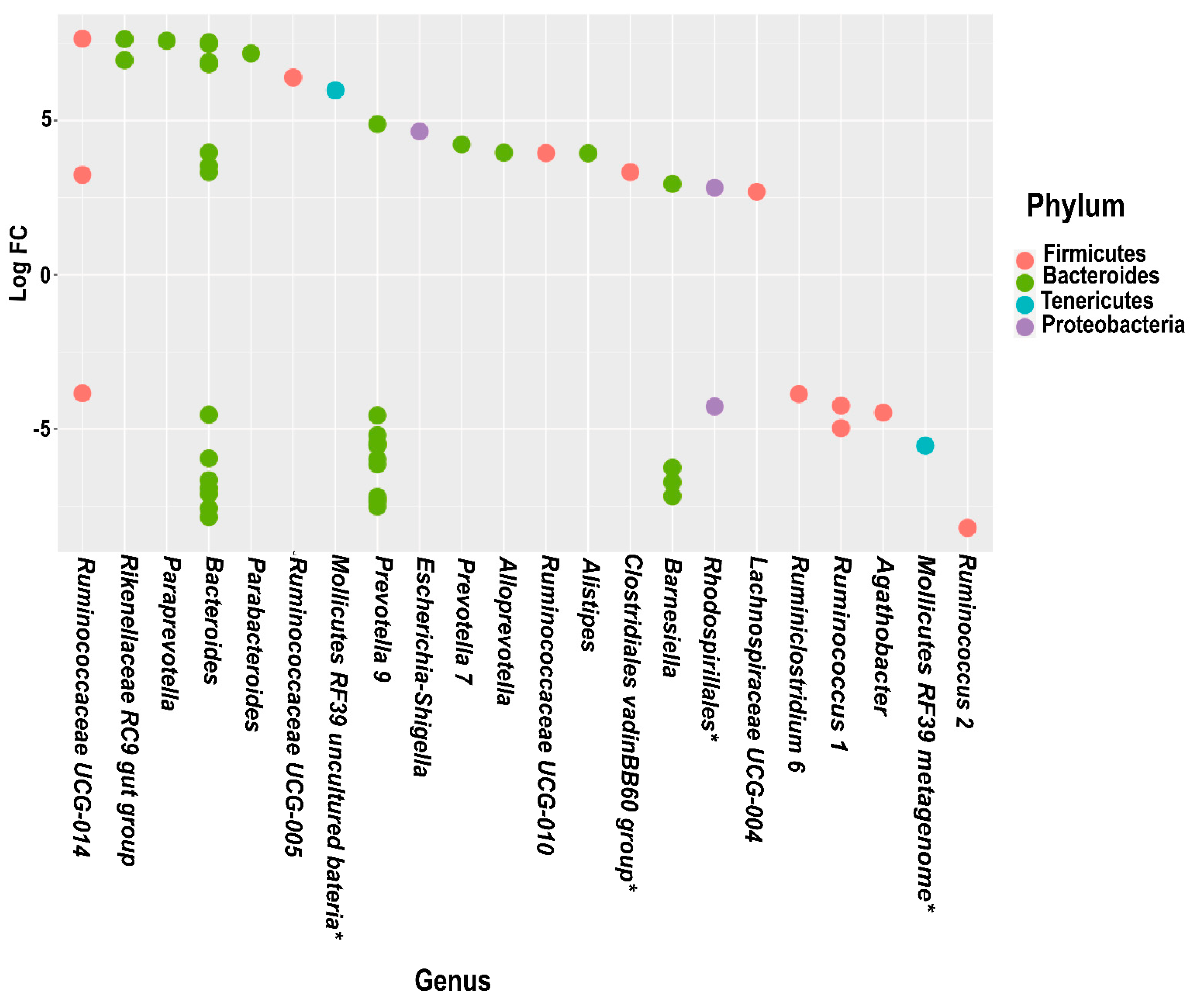
3.5. Differential Abundance Analysis
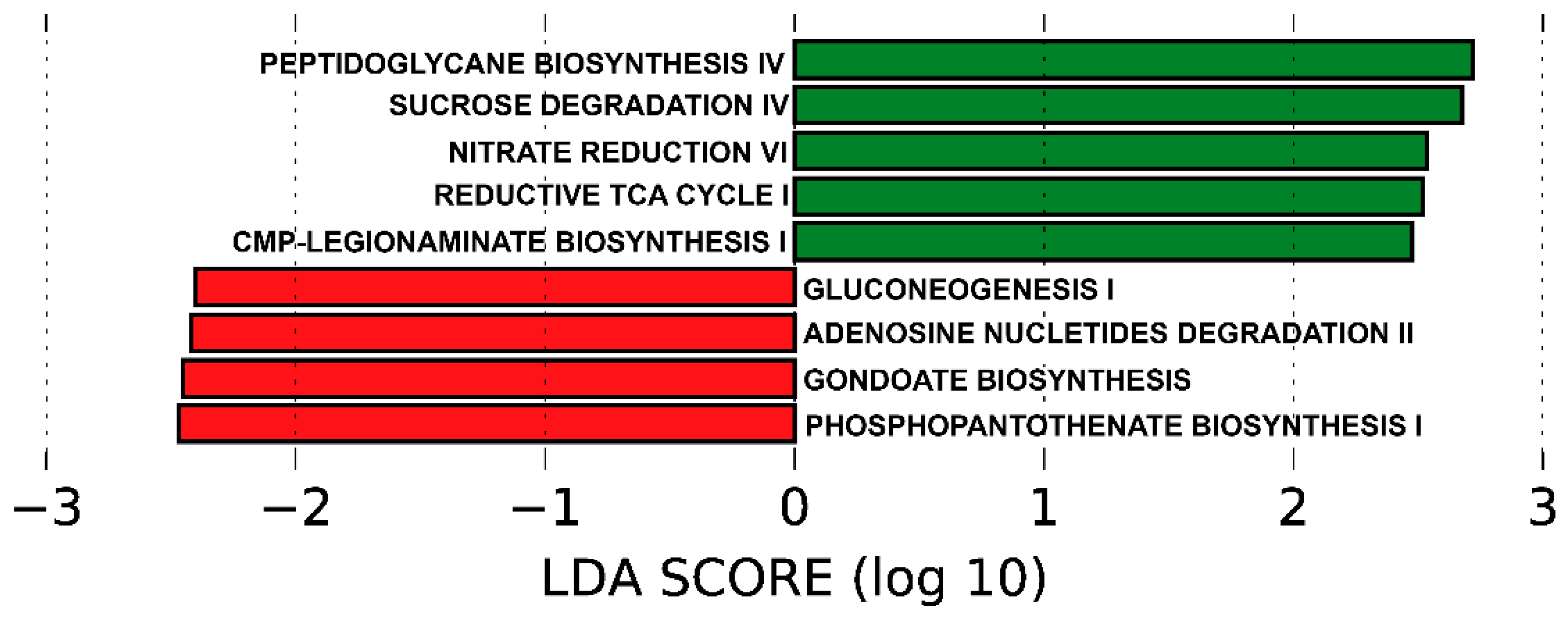
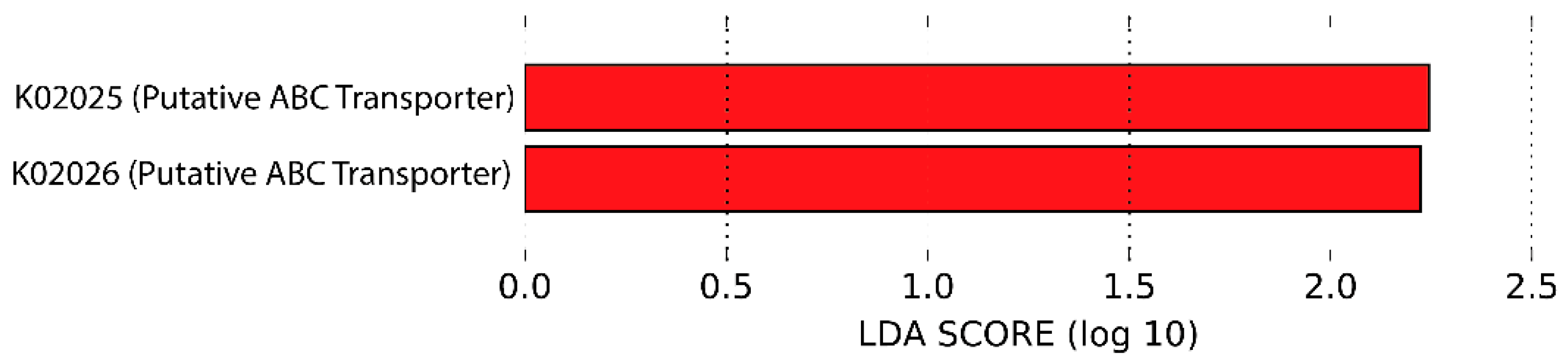
3.6. Predicted Functional Metagenome by PICRUSt
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Weeks, T.L.; Hazen, S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020, 127, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Bäckhed, F. Diet–microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Cerdá, B.; Pérez, M.; Pérez-Santiago, J.D.; Tornero-Aguilera, J.F.; González-Soltero, R.; Larrosa, M. Gut Microbiota Modification: Another Piece in the Puzzle of the Benefits of Physical Exercise in Health? Front. Physiol. 2016, 7, 51. [Google Scholar] [CrossRef]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbio. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 1–12. [Google Scholar] [CrossRef]
- Khachatryan, Z.A.; Ktsoyan, Z.A.; Manukyan, G.P.; Kelly, D.; Ghazaryan, K.A.; Aminov, R.I. Predominant Role of Host Genetics in Controlling the Composition of Gut Microbiota. PLoS ONE 2008, 3, e3064. [Google Scholar] [CrossRef]
- Li, E.; Hamm, C.M.; Gulati, A.S.; Sartor, R.B.; Chen, H.; Wu, X.; Zhang, T.; Rohlf, F.J.; Zhu, W.; Gu, C.; et al. Inflammatory Bowel Diseases Phenotype, C. difficile and NOD2 Genotype Are Associated with Shifts in Human Ileum Associated Microbial Composition. PLoS ONE 2012, 7, e26284. [Google Scholar] [CrossRef]
- Tong, M.; McHardy, I.; Ruegger, P.; Goudarzi, M.; Kashyap, P.C.; Haritunians, T.; Li, X.; Graeber, T.G.; Schwager, E.; Huttenhower, C.; et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn’s disease risk polymorphism. ISME J. 2014, 8, 2193–2206. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Silverberg, M.S.; Weersma, R.K.; Gevers, D.; Dijkstra, G.; Huang, H.; Tyler, A.D.; Van Sommeren, S.; Imhann, F.; Stempak, J.M.; et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef]
- Skogsberg, J.; Kannisto, K.; Roshani, L.; Gagné, E.; Hamsten, A.; Larsson, C.; Ehrenborg, E. Characterization of the human peroxisome proliferator activated receptor delta gene and its expression. Int. J. Mol. Med. 2000, 6, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Holst, D.; Luquet, S.; Nogueira, V.; Kristiansen, K.; Leverve, X.; Grimaldi, P.A. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2003, 1633, 43–50. [Google Scholar] [CrossRef]
- Russell, A.P.; Hesselink, M.K.C.; Lo, S.K.; Schrauwen, P. Regulation of metabolic transcriptional co-activators and transcription factors with acute exercise. FASEB J. 2005, 19, 986–988. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Olson, P.; Hevener, A.; Mehl, I.; Chong, L.-W.; Olefsky, J.M.; Gonzalez, F.J.; Ham, J.; Kang, H.; Peters, J.M.; et al. PPAR regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 2006, 103, 3444–3449. [Google Scholar] [CrossRef]
- Vänttinen, M.; Nuutila, P.; Kuulasmaa, T.; Pihlajamäki, J.; Hällsten, K.; Virtanen, K.A.; Lautamäki, R.; Peltoniemi, P.; Takala, T.; Viljanen, A.P.M.; et al. Single Nucleotide Polymorphisms in the Peroxisome Proliferator–Activated Receptor δ Gene Are Associated with Skeletal Muscle Glucose Uptake. Diabetes 2005, 54, 3587–3591. [Google Scholar] [CrossRef][Green Version]
- Andrulionyte, L.; Peltola, P.; Chiasson, J.-L.; Laakso, M.; STOP-NIDDM Study Group. Single Nucleotide Polymorphisms of PPARD in Combination with the Gly482Ser Substitution of PGC-1A and the Pro12Ala Substitution of PPARG2 Predict the Conversion from Impaired Glucose Tolerance to Type 2 Diabetes: The STOP-NIDDM Trial. Diabetes 2006, 55, 2148–2152. [Google Scholar] [CrossRef]
- Stefan, N.; Thamer, C.; Staiger, H.; Machicao, F.; Machann, J.; Schick, F.; Venter, C.; Niess, A.; Laakso, M.; Fritsche, A.; et al. Genetic Variations inPPARDandPPARGC1ADetermine Mitochondrial Function and Change in Aerobic Physical Fitness and Insulin Sensitivity during Lifestyle Intervention. J. Clin. Endocrinol. Metab. 2007, 92, 1827–1833. [Google Scholar] [CrossRef]
- Lee, M.Y.; Choi, R.; Kim, H.M.; Cho, E.J.; Kim, B.H.; Choi, Y.S.; Naowaboot, J.; Lee, E.Y.; Yang, Y.C.; Shin, J.Y.; et al. Peroxisome proliferator-activated receptor δ agonist attenuates hepatic steatosis by anti-inflammatory mechanism. Exp. Mol. Med. 2012, 44, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Rius-Pérez, S.; Torres-Cuevas, I.; Millán, I.; Ortega, Á.L.; Pérez, S. PGC-1α, Inflammation, and Oxidative Stress: An Integrative View in Metabolism. Oxid. Med. Cell. Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Ek, J.; Andersen, G.; Urhammer, S.A.; Gæde, P.; Drivsholm, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. Mutation analysis of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia 2001, 44, 2220–2226. [Google Scholar] [CrossRef]
- Hara, K.; Tobe, K.; Okada, T.; Kadowaki, H.; Akanuma, Y.; Ito, C.; Kimura, S. A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to Type II diabetes. Diabetologia 2002, 45, 740–743. [Google Scholar] [CrossRef]
- Ussar, S.; Griffin, N.W.; Bezy, O.; Fujisaka, S.; Vienberg, S.; Softic, S.; Deng, L.; Bry, L.; Gordon, J.I.; Kahn, C.R. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab. 2015, 22, 516–530. [Google Scholar] [CrossRef]
- Oh, H.Y.P.; Visvalingam, V.; Wahli, W. The PPAR–microbiota–metabolic organ trilogy to fine-tune physiology. FASEB J. 2019, 33, 9706–9730. [Google Scholar] [CrossRef]
- Hasan, A.U.; Rahman, A.; Kobori, H. Interactions between Host PPARs and Gut Microbiota in Health and Disease. Int. J. Mol. Sci. 2019, 20, 387. [Google Scholar] [CrossRef]
- Freedson, P.S.; Melanson, E.; Sirard, J. Calibration of the Computer Science and Applications, Inc. accelerometer. Med. Sci. Sports Exerc. 1998, 30, 777–781. [Google Scholar] [CrossRef]
- García-Villalba, R.; Bastida, J.A.G.; Conesa, M.T.G.; Tomas-Barberan, F.; Espín, J.C.; Larrosa, M. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 2012, 35, 1906–1913. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa FABIANPEDREGOSA, F.; Michel, V.; Grisel OLIVIERGRISEL, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; Cournapeau, D.; Pedregosa, F.; Varoquaux, G.; et al. Scikit-Learn: Machine Learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2013, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Kaehler, B.D.; Bokulich, N.; Mcdonald, D.; Knight, R.; Caporaso, J.; Gregory, C.J.; Huttley, G.A. Species-Level Microbial Sequence Classification Is Improved by Source-Environment Information. bioRxiv 2019, 406611. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Prior, S.L.; Clark, A.R.; Jones, D.A.; Bain, S.C.; Hurel, S.J.; Humphries, S.E.; Stephens, J.W. Association of the PGC-1? rs 8192678 Variant with Microalbuminuria in Subjects with Type 2 Diabetes Mellitus. Dis. Markers 2012, 32, 363–369. [Google Scholar] [CrossRef]
- Karaderi, T.; Drong, A.W.; Lindgren, C.M. Insights into the Genetic Susceptibility to Type 2 Diabetes from Genome-Wide Association Studies of Obesity-Related Traits. Curr. Diabetes Rep. 2015, 15, 83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lai, C.-Q.; Tucker, K.L.; Parnell, L.D.; Adiconis, X.; García-Bailo, B.; Griffith, J.; Meydani, M.; Ordovás, J.M. PPARGC1A Variation Associated with DNA Damage, Diabetes, and Cardiovascular Diseases: The Boston Puerto Rican Health Study. Diabetes 2007, 57, 809–816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lillioja, S.; Mott, D.M.; Spraul, M.; Ferraro, R.; Foley, J.E.; Ravussin, E.; Knowler, W.C.; Bennett, P.H.; Bogardus, C. Insulin Resistance and Insulin Secretory Dysfunction as Precursors of Non-Insulin-Dependent Diabetes Mellitus: Prospective Studies of Pima Indians. N. Engl. J. Med. 1993, 329, 1988–1992. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.C.; Warram, J.H.; Krolewski, A.S.; Soeldner, J.S.; Kahn, C.R.; Bergman, R.N. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: Results of a 25-year follow-up study. Lancet 1992, 340, 925–929. [Google Scholar] [CrossRef]
- Bloomgarden, Z.T. Insulin resistance: Current concepts. Clin. Ther. 1998, 20, 216–231. [Google Scholar] [CrossRef]
- Kim, J.A.; Wei, Y.; Sowers, J.R. Role of Mitochondrial Dysfunction in Insulin Resistance. Circ. Res. 2008, 102, 401–414. [Google Scholar] [CrossRef]
- Sesti, G. Pathophysiology of insulin resistance. Best Pract. Res. Clin. Endocrinol. Metab. 2006, 20, 665–679. [Google Scholar] [CrossRef]
- Petersen, K.F.; Shulman, G.I. Etiology of Insulin Resistance. Am. J. Med. 2006, 119, S10–S16. [Google Scholar] [CrossRef]
- Csép, K.; Szigeti, E.; Vitai, M.; Koranyi, L. The Ppargc1A-Gly482Ser Polymorphism (RS8192678) and the Metabolic Syndrome in a Central Romanian Population. Acta Endocrinol. 2017, 13, 161–167. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Y.; Wang, X.; Wu, X.; Zhu, X.; Li, J.; Ma, J.; Gu, H.F.; Liu, Y. Evaluation of the association between the PPARGC1A genetic polymorphisms and type 2 diabetes in Han Chinese population. Diabetes Res. Clin. Pract. 2009, 86, 168–172. [Google Scholar] [CrossRef]
- Bhat, A.; Koul, A.; Rai, E.; Sharma, S.; Dhar, M.K.; Bamezai, R.N.K. PGC-1α Thr394Thr and Gly482Ser variants are significantly associated with T2DM in two North Indian populations: A replicate case-control study. Qual. Life Res. 2007, 121, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Esterbauer, H.; Oberkofler, H.; Linnemayr, V.; Iglseder, B.; Hedegger, M.; Wolfsgruber, P.; Paulweber, B.; Fastner, G.; Krempler, F.; Patsch, W. Peroxisome Proliferator-Activated Receptor-γ Coactivator-1 Gene Locus: Associations with Obesity Indices in Middle-Aged Women. Diabetes 2002, 51, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Vohl, M.-C.; Houde, A.; Lebel, S.; Hould, F.-S.; Marceau, P. Effects of the peroxisome proliferator-activated receptor-γ co-activator-1 Gly482Ser variant on features of the metabolic syndrome. Mol. Genet. Metab. 2005, 86, 300–306. [Google Scholar] [CrossRef]
- Lacquemant, C.; Chikri, M.; Boutin, P.; Samson, C.; Froguel, P. No association between the G482S polymorphism of the proliferator-activated receptor-gamma coactivator-1 (PGC-1) gene and Type II diabetes in French Caucasians. Diabetologia 2002, 45, 602. [Google Scholar] [CrossRef]
- Stumvoll, M.; Fritsche, A.; T’Hart, L.M.; Machann, J.; Thamer, C.; Tschritter, O.; Van Haeften, T.W.; Jacob, S.; Dekker, J.M.; Maassen, J.A.; et al. The Gly482Ser Variant in the Peroxisome Proliferator-Activated Receptor γ Coactivator-1 is not Associated with Diabetes-Related Traits in Non-Diabetic German and Dutch Populations. Exp. Clin. Endocrinol. Diabetes 2004, 112, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Franson, J.; Grose, J.; Larson, K.; Bridgewater, L. Gut Microbiota Regulates the Interaction between Diet and Genetics to Influence Glucose Tolerance. Medicines 2021, 8, 34. [Google Scholar] [CrossRef]
- Pinna, N.K.; Anjana, R.M.; Saxena, S.; Dutta, A.; Gnanaprakash, V.; Rameshkumar, G.; Aswath, S.; Raghavan, S.; Rani, C.S.S.; Radha, V.; et al. Trans-ethnic gut microbial signatures of prediabetic subjects from India and Denmark. Genome Med. 2021, 13, 36. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, Q.; Ji, H.; Ning, J.; Li, C.; Zheng, H. Type 1 diabetes induces cognitive dysfunction in rats associated with alterations of the gut microbiome and metabolomes in serum and hippocampus. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 165541. [Google Scholar] [CrossRef]
- Kostic, A.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Kameyama, K.; Itoh, K. Intestinal Colonization by a Lachnospiraceae Bacterium Contributes to the Development of Diabetes in Obese Mice. Microbes Environ. 2014, 29, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.J.; Abratt, V.R. Sucrose utilisation in bacteria: Genetic organisation and regulation. Appl. Microbiol. Biotechnol. 2005, 67, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Hamaguchi, M.; Kaji, A.; Sakai, R.; Osaka, T.; Inoue, R.; Kashiwagi, S.; Mizushima, K.; Uchiyama, K.; Takagi, T.; et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J. Diabetes Investig. 2020, 11, 1623–1634. [Google Scholar] [CrossRef]
- Huergo, L.; Dixon, R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol. Mol. Biol. Rev. 2015, 79, 419–435. [Google Scholar] [CrossRef]
- Arora, A.; Behl, T.; Sehgal, A.; Singh, S.; Sharma, N.; Bhatia, S.; Sobarzo-Sanchez, E.; Bungau, S. Unravelling the involvement of gut microbiota in type 2 diabetes mellitus. Life Sci. 2021, 273, 119311. [Google Scholar] [CrossRef]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Hou, Y.-P.; He, Q.-Q.; Ouyang, H.-M.; Peng, H.-S.; Wang, Q.; Li, J.; Lv, X.-F.; Zheng, Y.-N.; Li, S.-C.; Liu, H.-L.; et al. Human Gut Microbiota Associated with Obesity in Chinese Children and Adolescents. BioMed Res. Int. 2017, 2017, 7585989. [Google Scholar] [CrossRef]
- Behl, T.; Sehgal, A.; Grover, M.; Singh, S.; Sharma, N.; Bhatia, S.; Al-Harrasi, A.; Aleya, L.; Bungau, S. Uncurtaining the pivotal role of ABC transporters in diabetes mellitus. Environ. Sci. Pollut. Res. 2021, 28, 41533–41551. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Candela, M.; Biagi, E.; Soverini, M.; Consolandi, C.; Quercia, S.; Severgnini, M.; Peano, C.; Turroni, S.; Rampelli, S.; Pozzilli, P.; et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br. J. Nutr. 2016, 116, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Rahnavard, G.; et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, J.; Cao, Y.; Zhang, G.; Chen, Y.; Zhong, J.; Huang, W.; Zeng, J.; Wu, P. Relationship between alterations of urinary microbiota and cultured negative lower urinary tract symptoms in female type 2 diabetes patients. BMC Urol. 2019, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Li, L.; Li, L.; Liu, Y.; Ren, Q.; Shi, M.; Liu, J.; Jiang, J.; Ma, H.; Huang, Z.; et al. Understanding the gut–kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: An analysis of the gut microbiota composition. Geol. Rundsch. 2019, 56, 581–592. [Google Scholar] [CrossRef]
| PPARD (rs 2267668) | PPARGC1A (rs 8192678) | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype Frequency | Expected Frequency * | Allelic Frequencies | Genotype Frequency | Expected Frequency ** | Allelic Frequencies | |||
| AA | 0.62 | 0.67 | Allele A | 0.79 | CC 0.934 | 0.445 | Allele C | 0.967 |
| AG | 0.34 | 0.29 | Allele G | 0.21 | CT 0.066 | 0.444 | Allele T | 0.033 |
| GG | 0.039 | 0.035 | TT 0 | 0.111 | ||||
| PPARD-1 | PPARD-2 | p | PPARGC1A-1 | PPARGC1A-2 | p | |
|---|---|---|---|---|---|---|
| Sex (n/%) | 23/50 M 23/50 W | 15/55.6 M 12/44.4 W | 0.796 | 36/51.4 M 34/48.6 W | 4/66.7 M 2/33.3 W | 0.677 * |
| Age (years) | 33.73 ± 7.40 | 33.73 ± 8.06 | 1.00 | 33.26 ± 7.87 | 36.83 ± 2.04 | 0.27 |
| Body mass (kg) | 69.25 ± 13.05 | 70.27 ± 12.20 | 0.75 | 69.22 ± 12.93 | 73.75 ± 8.11 | 0.40 |
| BMI (kg/m2) | 24.21 ± 3.61 | 23.77 ± 3.12 | 0.61 | 23.94 ± 3.53 | 25.40 ± 1.42 | 0.32 |
| BFP (%) | 26.07 ± 7.48 | 27.82 ± 8.56 | 0.39 | 27.21 ± 7.90 | 23.22 ± 6.26 | 0.28 |
| BFM (kg) | 17.36 ± 6.59 | 18.95 ± 6.02 | 0.33 | 18.15 ± 6.47 | 17.03 ± 4.90 | 0.71 |
| VAT (g) | 332.98 ± 192.11 | 356.20 ± 181.26 | 0.63 | 343.53 ± 179.66 | 368.80 ± 257.95 | 0.77 |
| AI (kg/m2) | 6.06 ± 2.12 | 6.4 ± 2.32 | 0.45 | 6.32 ± 2.20 | 5.76 ± 1.69 | 0.58 |
| MMI (kg/m2) | 16.05 ± 2.21 | 15.77 ± 2.57 | 0.63 | 15.77 ± 2.36 | 17.98 ± 1.41 | 0.04 ** |
| AppMMI (kg/m2) | 7.17 ± 1.29 | 6.99 ± 1.42 | 0.60 | 7.02 ± 1.35 | 8.08 ± 0.74 | 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bailén, M.; Tabone, M.; Bressa, C.; Lominchar, M.G.M.; Larrosa, M.; González-Soltero, R. Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population. Genes 2022, 13, 289. https://doi.org/10.3390/genes13020289
Bailén M, Tabone M, Bressa C, Lominchar MGM, Larrosa M, González-Soltero R. Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population. Genes. 2022; 13(2):289. https://doi.org/10.3390/genes13020289
Chicago/Turabian StyleBailén, María, Mariangela Tabone, Carlo Bressa, María Gregoria Montalvo Lominchar, Mar Larrosa, and Rocío González-Soltero. 2022. "Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population" Genes 13, no. 2: 289. https://doi.org/10.3390/genes13020289
APA StyleBailén, M., Tabone, M., Bressa, C., Lominchar, M. G. M., Larrosa, M., & González-Soltero, R. (2022). Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population. Genes, 13(2), 289. https://doi.org/10.3390/genes13020289










