The Role of the Disrupted Podosome Adaptor Protein (SH3PXD2B) in Frank–Ter Haar Syndrome
Abstract
1. Introduction
2. Patients and Methods
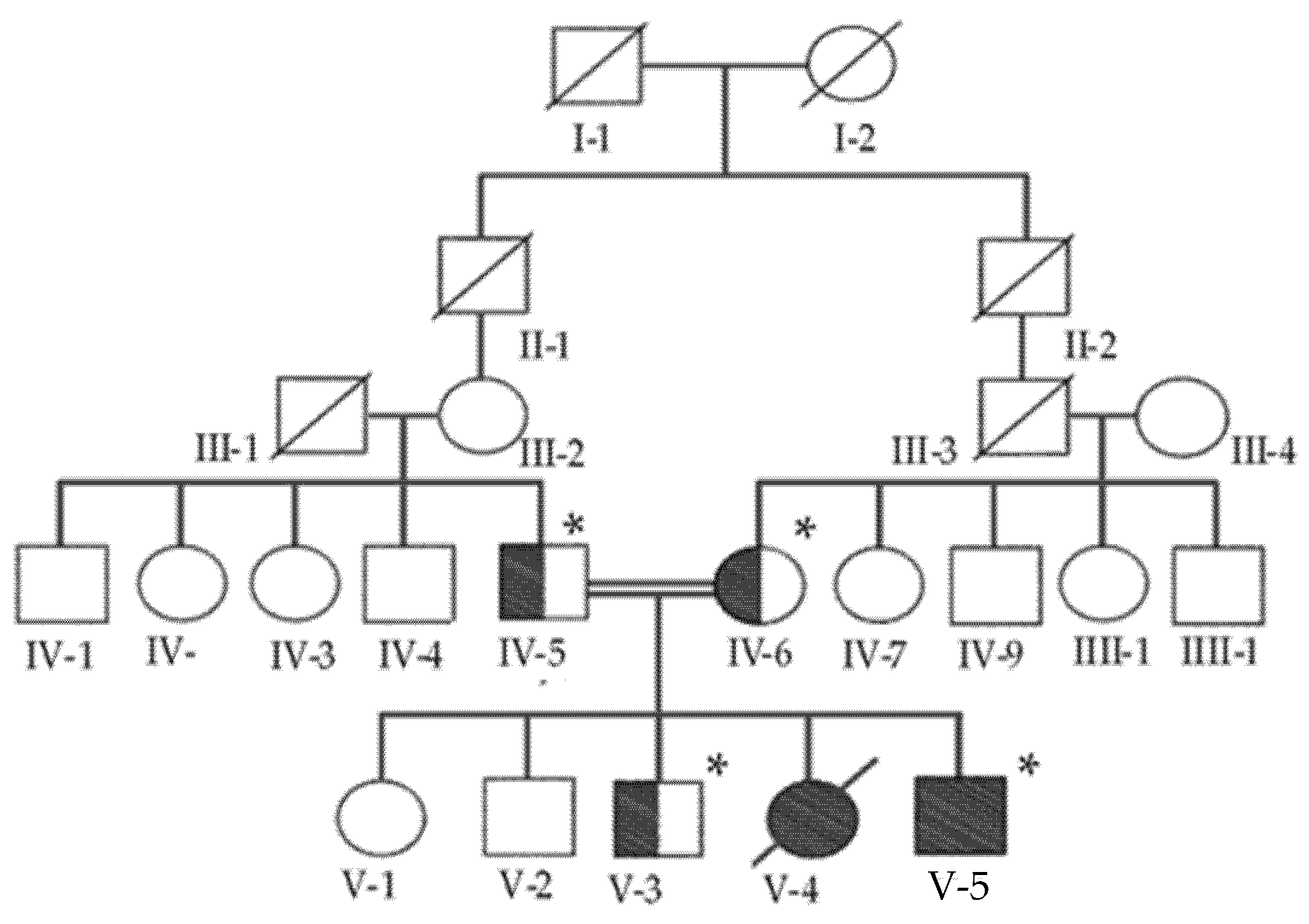
2.1. Patient Description and Ethical Considerations
2.2. Genomic DNA Extraction
2.3. Whole-Exome Sequencing (WES)
2.4. Variant Annotation and Filtering
2.5. Sanger Sequencing
3. Results
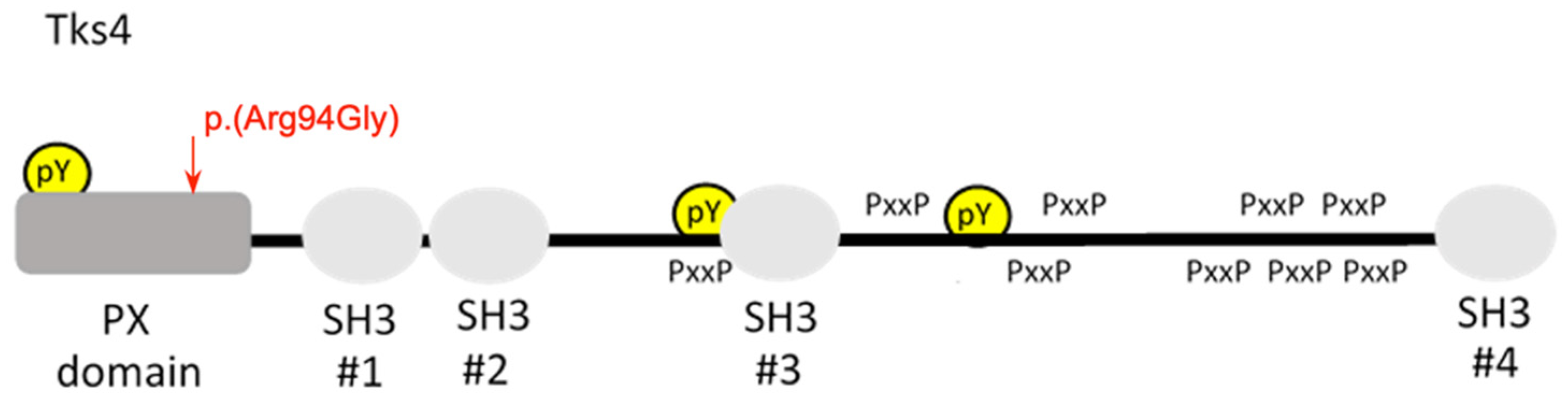
Identification of Missense Mutation in SH3BPXD2B
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FTHS | Frank–Ter Haar syndrome |
| IAA | Interrupted aortic arch |
| PDA | Patent ductus arteriosus |
| VSD | Ventricle septal defect |
| CHD | Congenital heart disease |
| IVH | Intraventricular hemorrhage |
| GERD | Gastroesophageal reflux disease |
| CoA | Coarctation of the aorta |
| MVP | Mitral valve prolapse |
References
- Durand, B.; Stoetzel, C.; Schaefer, E.; Calmels, N.; Scheidecker, S.; Kempf, N.; De Melo, C.; Guilbert, A.-S.; Timbolschi, D.; Donato, L.; et al. A severe case of Frank-ter Haar syndrome and literature review: Further delineation of the phenotypical spectrum. Eur. J. Med. Genet. 2020, 63, 103857. [Google Scholar] [CrossRef] [PubMed]
- Ratukondla, B.; Prakash, S.; Reddy, S.; Puthuran, G.V.; Kannan, N.B.; Pillai, M.R. A Rare Case Report of Frank Ter Haar Syndrome in a Sibling Pair Presenting With Congenital Glaucoma. J. Glaucoma 2019, 29, 236–238. [Google Scholar] [CrossRef] [PubMed]
- ter Haar, B.; Hamel, B.; Hendriks, J.; de Jager, J.; Opitz, J.M. Melnick-Needles syndrome: Indication for an autosomal recessive form. Am. J. Med. Genet. 1982, 13, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Frank, Y.; Ziprkowski, M.; Romano, A.; Stein, R.; Katznelson, M.B.; Cohen, B.; Goodman, R.M. Megalocornea associ-ated with multiple skeletal anomalies: A new genetic syndrome? J. Genet. Hum. 1973, 21, 67–72. [Google Scholar] [PubMed]
- Bendon, C.L.; Fenwick, A.L.; Hurst, J.; Nürnberg, G.; Nürnberg, P.; Wall, S.; Wilkie, A.O.; Johnson, D. Frank-ter Haar syndrome associated with sagittal craniosynostosis and raised intracranial pressure. BMC Med. Genet. 2012, 13, 104. [Google Scholar] [CrossRef]
- Iqbal, Z.; Cejudo-Martin, P.; de Brouwer, A.; van der Zwaag, B.; Ruiz-Lozano, P.; Scimia, M.C.; Lindsey, J.D.; Weinreb, R.; Albrecht, B.; Megarbane, A.; et al. Disruption of the Podosome Adaptor Protein TKS4 (SH3PXD2B) Causes the Skeletal Dysplasia, Eye, and Cardiac Abnormalities of Frank-Ter Haar Syndrome. Am. J. Hum. Genet. 2010, 86, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Maddirevula, S.; Alsahli, S.; Alhabeeb, L.; Patel, N.; Alzahrani, F.; Shamseldin, H.E.; Anazi, S.; Ewida, N.; Alsaif, H.S.; Mohamed, J.Y.; et al. Expanding the phenome and variome of skeletal dysplasia. Genet. Med. Off. J. Am. Coll. Med. Genet. 2018, 20, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Türkyilmaz, A.; Sager, S.G.; Topcu, B.; Kaplan, A.T.; Günbey, H.P.; Akin, Y. Novel SH3PXD2B variant identified by whole-exome sequencing in a Turkish newborn with Frank–Ter Haar Syndrome. Clin. Dysmorphol. 2021, 31, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.M.; Kayserili, H.; Lam, J.; Apak, M.Y.; Hennekam, R.C. Further delineation of Frank-ter Haar syndrome. Am. J. Med. Genet. 2004, 131, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.R.; Sunley, J.; Smith, K.R.; Pope, K.; Bromhead, C.J.; Fitzpatrick, E.; Di Rocco, M.; van Steensel, M.; Coman, D.; Leventer, R.J.; et al. Mutations in SH3PXD2B cause Borrone dermato-cardio-skeletal syndrome. Eur. J. Hum. Genet. 2013, 22, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Bögel, G.; Gujdár, A.; Geiszt, M.; Lányi, A.; Fekete, A.; Sipeki, S.; Downward, J.; Buday, L. Frank-ter Haar Syndrome Protein Tks4 Regulates Epidermal Growth Factor-dependent Cell Migration. J. Biol. Chem. 2012, 287, 31321–31329. [Google Scholar] [CrossRef] [PubMed]
- Borrone, C.; Di Rocco, M.; Crovato, F.; Camera, G.; Gambini, C. New multisystemic disorder involving heart valves, skin, bones, and joints in two brothers. Am. J. Med. Genet. 1993, 46, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Buschman, M.D.; Bromann, P.A.; Cejudo-Martin, P.; Wen, F.; Pass, I.; Courtneidge, S.A. The Novel Adaptor Protein Tks4 (SH3PXD2B) Is Required for Functional Podosome Formation. Mol. Biol. Cell 2009, 20, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Thedens, D.R.; Chang, B.; Harris, B.S.; Zheng, Q.Y.; Johnson, K.R.; Donahue, L.R.; Anderson, M.G. The podosomal-adaptor protein SH3PXD2B is essential for normal postnatal development. Mamm. Genome 2009, 20, 462–475. [Google Scholar] [CrossRef][Green Version]
- Mégarbané, A.; Tomey, K.; Wakim, G. Congenital glaucoma, limb deformities, skeletal dysplasia, and facial anomalies: Report of another family. Am. J. Med. Genet. 1997, 73, 67–71. [Google Scholar] [CrossRef]
- Nagase, T.; Kikuno, R.; Ishikawa, K.-I.; Hirosawa, M.; Ohara, O. Prediction of the Coding Sequences of Unidentified Human Genes. XVII. The Complete Sequences of 100 New cDNA Clones from Brain Which Code for Large Proteins in vitro. DNA Res. 2000, 7, 143–150. [Google Scholar] [CrossRef]

| Features | Patient V-5 |
|---|---|
| Age | 0.33 years |
| Gender | Male |
| Weight | 4.78 kg |
| Height | 55 cm |
| Consanguinity | Yes |
| Cardiovascular pathology | CHD in the form of severe CoA versus I.A.A., small aortic arch, P.D.A., V.S.D., small parachute-like mitral valve |
| Clinical diagnosis | Frank–Ter Haar syndrome |
| Neurological abnormalities | I.V.H., mild superior cerebellar vermis atrophy, wide fontanel, bilateral glaucoma |
| Motor development | Limb abnormalities, dysmorphic features |
| Other | Low-set ears, congenital glaucoma, dysmorphic features, GERD, talipes, choroid plexus cysts, portal vein thrombosis, gallbladder stone |
| SH3PXD2B Variants | Mutation | Inheritance | Number and Gender of Patients | Heritage of Patient | Clinical Diagnosis | Reference |
|---|---|---|---|---|---|---|
| c.280C>G:p. (Arg94Gly) | Missense point mutation | Autosomal recessive | 1 Male | Saudi | Wide fontanel, proptosis, low-set ears, talipes, hypertelorism, synophrys, lower limb abnormalities, right finger anomalies, small head, valve anomalies, severe coarctation, P.D.A., V.S.D., mild superior cerebellar vermis atrophy, congenital bilateral glaucoma | This study |
| Novel missense point mutation (first finding) | Autosomal recessive | 1 Male | Arab | Megalocornea, congenital cataract, congenital glaucoma, failure to thrive, GDD. The patient’s parents are consanguineous with a family history of a similar condition in 3 deceased cousins with a similar phenotype | [7] | |
| c.578delG, p.S193fs*48 | Novel frameshift variant | Autosomal recessive | 1 Female | Turkish | Prominent forehead, brachycephaly, wide anterior fontanel, proptosis, hypertelorism, full cheeks, anteverted nostrils, broad mouth, kyphoscoliosis, short hands, camptodactyly, toe anomalies, clubfeet, congenital glaucoma, megalocornea | [8] |
| c.969delG, p(Arg324Glyfs*19) | Variant altering the formation of podosomes and invadopodia | Autosomal recessive | 1 Female | Armenian | IUGR, hypotonia, congenital glaucoma, caudal appendix, scoliosis, camptodactyly, VSD, corpus callosum abnormality, craniofacial defects, buphthalmos, respiratory failure | [1] |
| chromosome 5q35.1 | Loss of function variant (exon 13 deletion) | Autosomal recessive | 2 Males and 1 Female | Pakistani | Prominent forehead, hypertelorism, brachycephaly, prominent ears, flat nasal bridge, full cheeks, hypoplasia of the teeth, broad mouth | [5] |
| chromosome 5q35.1 | Insertion at c.147insT, nonsense variant Deletion c.969delG, nonsense variant (p.G323fsX19) Missense variant c.129C>T (p.R43W) Splice-altering variant c.76-2A>C | Autosomal recessive | 1 Male | Dutch | Motor retardation, brachycephaly, prominent forehead, hypertelorism, wide anterior fontanels, prominent eyes, congenital glaucoma, large cornea, full cheeks, broad mouth, prominent coccyx, shorthands, megalocornea, clubfeet, anteverted nostrils, thoracolumbar kyphosis, heart murmur, flexion deformity of fingers | [6] |
| 3 Males | N.R. | [3] | ||||
| 1 Male and 1 Female | N.R. | [3] | ||||
| 3 Males | Arab | [6] | ||||
| 1 Female | Turkish | [9] | ||||
| 1 Male | Turkish | [6] | ||||
| 1 Male | Israeli | [6] | ||||
| c.1188+1773_2733+6592del | Complete loss of SH3PXD2B 12,583 bp deletion | Autosomal recessive | 2 Males | Italian | Thick skin, AC, coarse facial features, osteolysis, gingival hyperplasia, brachydactyly, camptodactyly, MVP, heart failure | [10] |
| c.401+1G-A | Complete loss of SH3PXD2B Splice-altering variant (Glu134GlufsTer1) | Autosomal recessive | 1 Male | Australian | Coarse facial features, brachydactyly, pachyermodactyly, MCP, MVP leading to acute congestive cardiac failure, respiratory failure | [10] |
| c.969delG; p. (Arg324Glyfs*19) | 1 bp deletion, frameshift variant | Autosomal recessive | 1 Female | Armenian | G.D.D., dysmorphic facial features, brachycephaly, prominent subocular folds, bilateral buphthalmos with megalocornea, bilateral congenital glaucoma, hip dysplasia | [1] |
| c.250C>T(p.R84*) | Nonsense substitution is predicted to cause premature truncation of the protein coded by the SH3PXD2B gene | Autosomal recessive | 2 Males | South Indian | Dysmorphic facial features, brachycephaly, pectus carinatum, kyphoscoliosis, megalocornea, MCP, congenital talipes equinovarus, prominent spine scapula | [2] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massadeh, S.; Alhabshan, F.; AlSudairi, H.N.; Alkwai, S.; Alsuwailm, M.; Kabbani, M.S.; Chaikhouni, F.; Alaamery, M. The Role of the Disrupted Podosome Adaptor Protein (SH3PXD2B) in Frank–Ter Haar Syndrome. Genes 2022, 13, 236. https://doi.org/10.3390/genes13020236
Massadeh S, Alhabshan F, AlSudairi HN, Alkwai S, Alsuwailm M, Kabbani MS, Chaikhouni F, Alaamery M. The Role of the Disrupted Podosome Adaptor Protein (SH3PXD2B) in Frank–Ter Haar Syndrome. Genes. 2022; 13(2):236. https://doi.org/10.3390/genes13020236
Chicago/Turabian StyleMassadeh, Salam, Fahad Alhabshan, Hadeel N. AlSudairi, Sarah Alkwai, Moneera Alsuwailm, Mohamed S. Kabbani, Farah Chaikhouni, and Manal Alaamery. 2022. "The Role of the Disrupted Podosome Adaptor Protein (SH3PXD2B) in Frank–Ter Haar Syndrome" Genes 13, no. 2: 236. https://doi.org/10.3390/genes13020236
APA StyleMassadeh, S., Alhabshan, F., AlSudairi, H. N., Alkwai, S., Alsuwailm, M., Kabbani, M. S., Chaikhouni, F., & Alaamery, M. (2022). The Role of the Disrupted Podosome Adaptor Protein (SH3PXD2B) in Frank–Ter Haar Syndrome. Genes, 13(2), 236. https://doi.org/10.3390/genes13020236






