Transcriptomic Analysis of MSTN Knockout in the Early Differentiation of Chicken Fetal Myoblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture
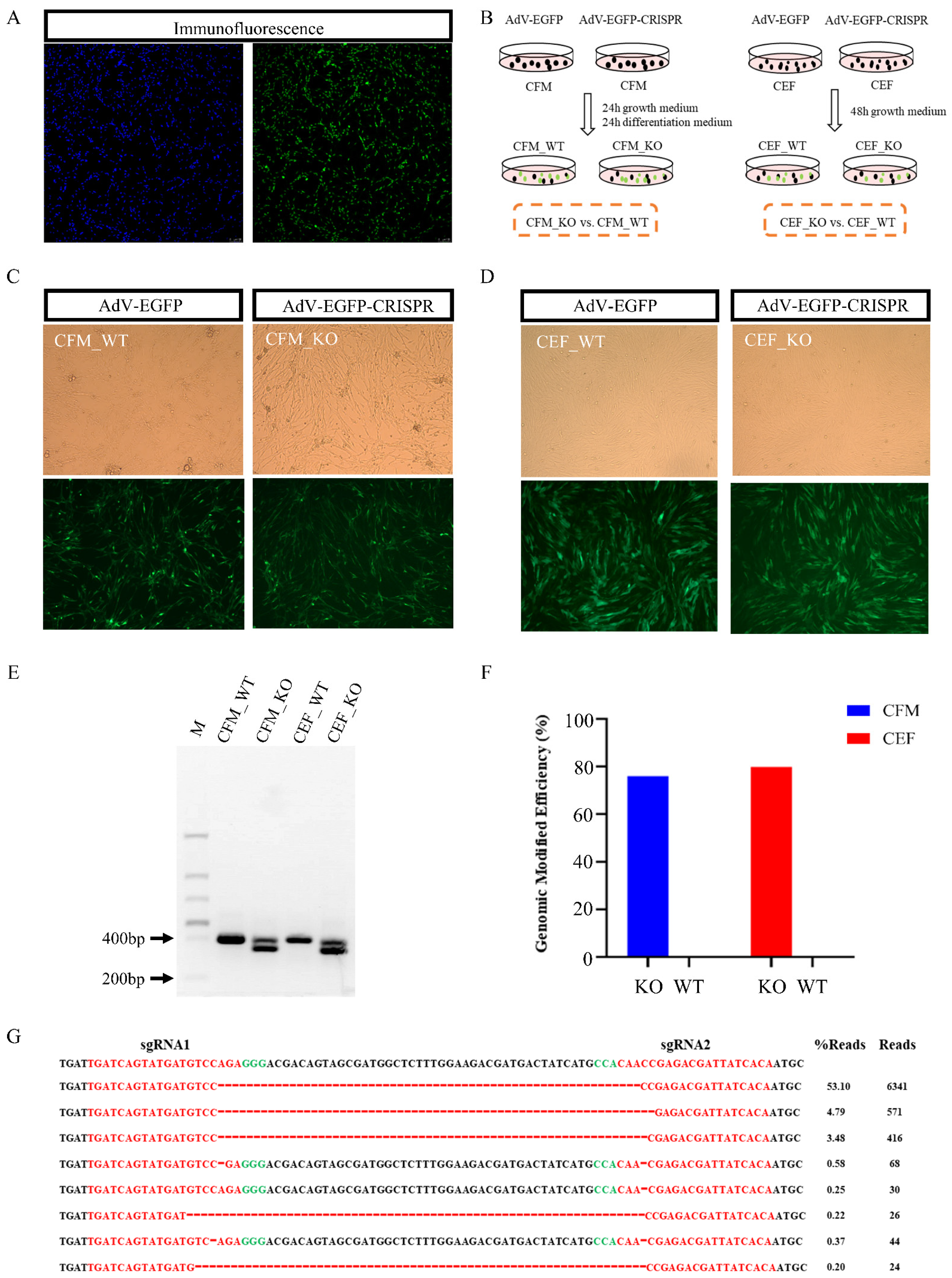
2.3. Immunofluorescence
2.4. Cell Transduction
2.5. Detection of Editing Effect and Efficiency by Amplicon Sequencing
2.6. Detection of Gene Expression by Transcriptome Sequencing
2.7. Principal Component Analysis (PCA) and Differentially Expressed Gene (DEG) Analysis
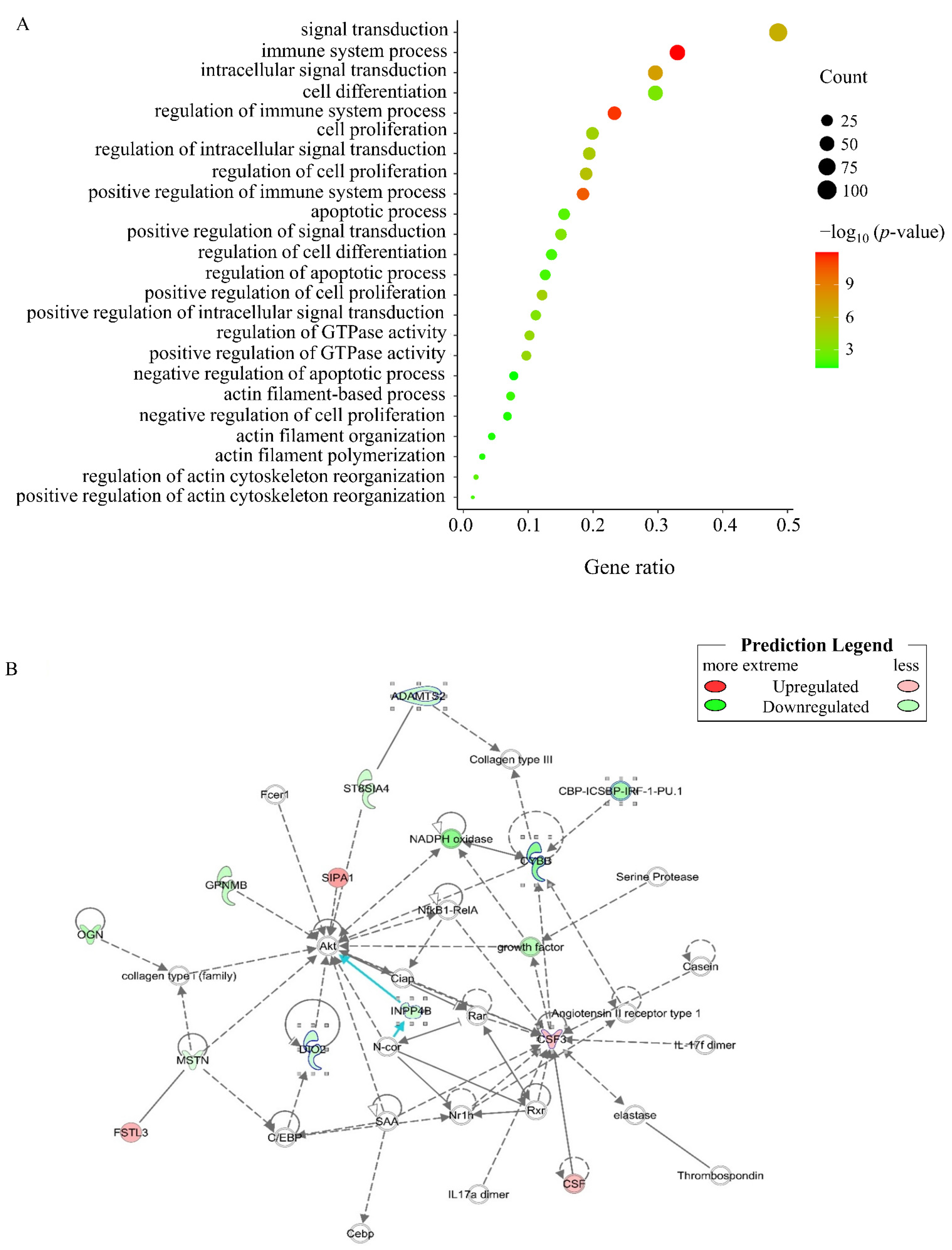
2.8. Enrichment Analysis of Differentially Expressed Genes (DEGs)
3. Results
3.1. Generation of CRISPR/Cas9-Mediated MSTN Knockout in Chicken Cells
3.2. Differentially Expressed Genes (DEGs) between KO and WT CFMs
3.3. Enrichment Analysis of DEGs
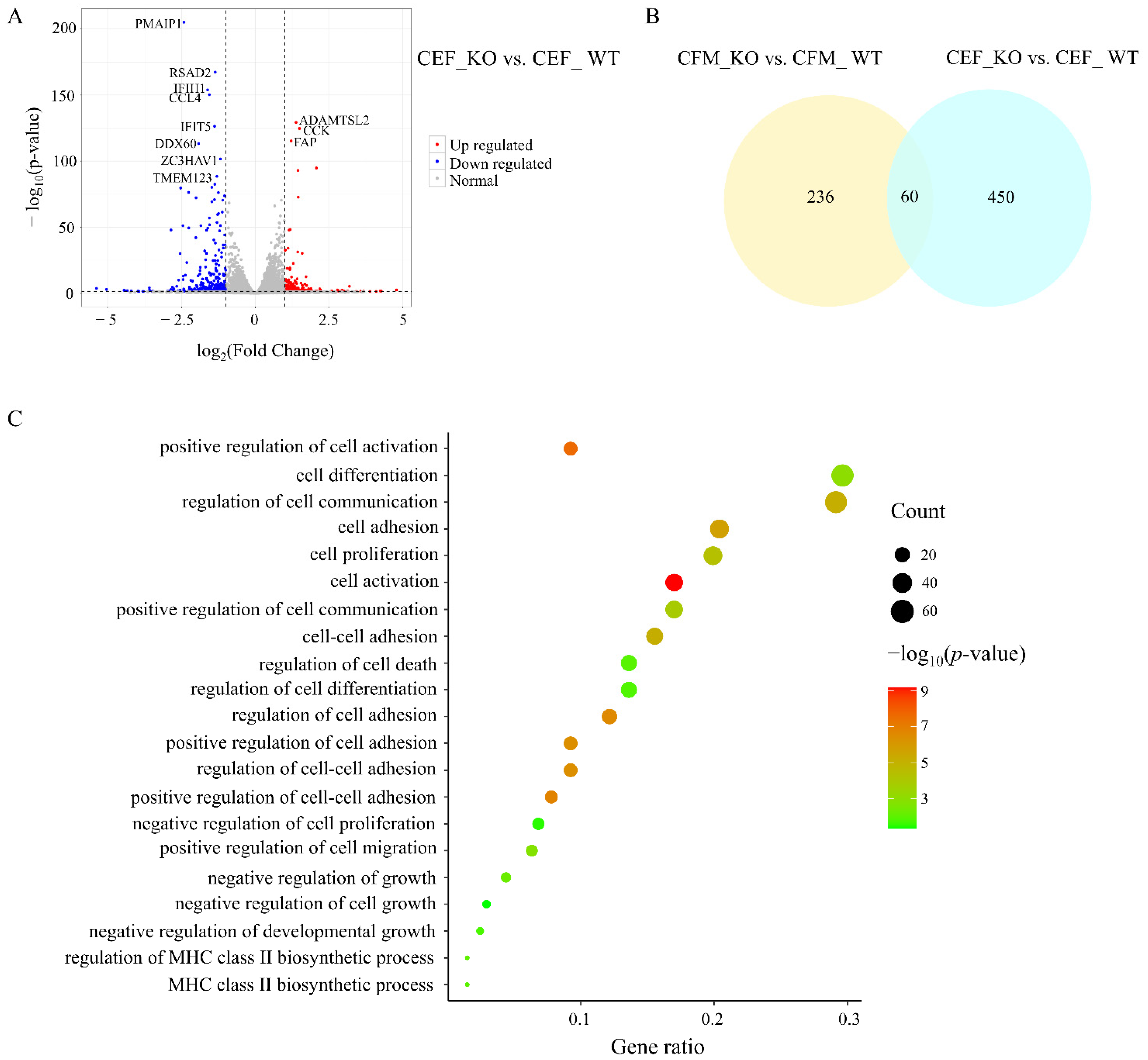
3.4. Comparison of Transcriptome Changes after MSTN KO in CFMs and CEFs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doran, T.J. Advances in genetic engineering of the avian genome: “Realising the promise”. Transgenic Res. 2016, 25, 110–111. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Lee, J.H.; Park, B.C.; Park, T.S. Myotube differentiation in clustered regularly interspaced short palindromic repeat/Cas9-mediated MyoD knockout quail myoblast cells. Asian-Australas. J. Anim. Sci. 2017, 30, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef] [PubMed]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–916. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. Super-muscly pigs created by small genetic tweak. Nature 2015, 523, 13–14. [Google Scholar] [CrossRef]
- Kim, Y.S.; Bobbili, N.K.; Paek, K.S.; Jin, H.J. Production of a monoclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult. Sci. 2006, 85, 1062–1071. [Google Scholar] [CrossRef]
- Kim, G.D.; Lee, J.H.; Song, S.M.; Kim, S.W.; Han, J.S.; Shin, S.P.; Park, B.C.; Park, T.S. Generation of myostatin-knockout chickens mediated by D10A-Cas9 nickase. Faseb J. 2020, 34, 5688–5696. [Google Scholar] [CrossRef]
- He, Z.Y.; Zhang, T.; Jiang, L.; Zhou, M.Y.; Wu, D.J.; Mei, J.Y.; Cheng, Y. Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Schuelke, M.; Wagner, K.R.; Stolz, L.E.; Hubner, C.; Riebel, T.; Komen, W.; Braun, T.; Tobin, J.F.; Lee, S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 350, 2682–2688. [Google Scholar] [CrossRef]
- Yang, W.; Chen, Y.; Zhang, Y.; Wang, X.; Yang, N.; Zhu, D. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res. 2006, 66, 1320–1326. [Google Scholar] [CrossRef]
- Elkasrawy, M.N.; Hamrick, M.W. Myostatin (GDF-8) as a key factor linking muscle mass and bone structure. J. Musculoskel. Neuron. Interact. 2010, 10, 56–63. [Google Scholar]
- Elkina, Y.; von Haehling, S.; Anker, S.D.; Springer, J. The role of myostatin in muscle wasting: An overview. J. Cachexia Sarcopeni. Muscle 2011, 2, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Paswan, C.; Bhattacharya, T.; Nagaraj, C.; Chaterjee, R.; Jayashankar, M. SNPs in minimal promoter of myostatin (GDF-8) gene and its association with body weight in broiler chicken. J. Appl. Anim. Res. 2014, 42, 304–309. [Google Scholar] [CrossRef][Green Version]
- Zhang, G.; Zhao, X.; Wang, J.; Ding, F.; Zhang, L. Effect of an exon 1 mutation in the myostatin gene on the growth traits of the Bian chicken. Anim. Genet. 2012, 43, 458–459. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.H.; Lee, K. Current Approaches and Applications in Avian Genome Editing. Int. J. Mol. Sci. 2020, 21, 3937. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Zhang, G.X.; Wu, P.F.; Duan, L.; Li, G.H.; Liu, Q.H.; Wang, J.Y. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes 2018, 9, 34. [Google Scholar] [CrossRef]
- Huang, P.; Pang, D.; Wang, K.; Xu, A.; Yao, C.; Li, M.; You, W.; Wang, Q.; Yu, H. The Possible Role of Complete Loss of Myostatin in Limiting Excessive Proliferation of Muscle Cells (C2C12) via Activation of MicroRNAs. Int. J. Mol. Sci. 2019, 20, 643. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, J.H.; Han, J.S.; Shin, S.P.; Park, T.S. Muscle differentiation induced by p53 signaling pathway-related genes in myostatin-knockout quail myoblasts. Mol. Biol. Rep. 2020, 47, 9531–9540. [Google Scholar] [CrossRef]
- Sato, F.; Kurokawa, M.; Yamauchi, N.; Hattori, M. Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. Am. J. Physiol.-Cell Physiol. 2006, 291, C538–C545. [Google Scholar] [CrossRef]
- Yi, X.; Tao, Y.; Lin, X.; Dai, Y.; Yang, T.L.; Yue, X.J.; Jiang, X.J.; Li, X.Y.; Jiang, D.S.; Andrade, K.C.; et al. Histone methyltransferase Setd2 is critical for the proliferation and differentiation of myoblasts. Bba-Mol. Cell Res. 2017, 1864, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Ringnér, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Vishal, K.; Lovato, T.L.; Bragg, C.; Chechenova, M.B.; Cripps, R.M. FGF signaling promotes myoblast proliferation through activation of wingless signaling. Dev. Biol. 2020, 464, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.K.; Levin, J.B.; Hamilton, A.M.; Borodinsky, L.N. Calcium signaling in skeletal muscle development, maintenance and regeneration. Cell Calcium 2016, 59, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Li, Y.L.; Wu, Y.N.; Wang, L.Y.; Wang, X.N.; Du, J. Interleukin-6/Signal Transducer and Activator of Transcription 3 (STAT3) Pathway Is Essential for Macrophage Infiltration and Myoblast Proliferation during Muscle Regeneration. J. Biol. Chem. 2013, 288, 1489–1499. [Google Scholar] [CrossRef]
- Wu, P.F.; Dai, G.J.; Chen, F.X.; Chen, L.; Zhang, T.; Xie, K.Z.; Wang, J.Y.; Zhan, G.X. Transcriptome profile analysis of leg muscle tissues between slow- and fast-growing chickens. PLoS ONE 2018, 13, e0206131. [Google Scholar] [CrossRef]
- Ayuso, M.; Fernandez, A.; Nunez, Y.; Benitez, R.; Isabel, B.; Barragan, C.; Fernandez, A.I.; Rey, A.I.; Medrano, J.F.; Canovas, A.; et al. Comparative Analysis of Muscle Transcriptome between Pig Genotypes Identifies Genes and Regulatory Mechanisms Associated to Growth, Fatness and Metabolism. PLoS ONE 2015, 10, e0145162. [Google Scholar] [CrossRef]
- Parsons, S.A.; Millay, D.P.; Sargent, M.A.; McNally, E.M.; Molkentin, J.D. Age-dependent effect of myostatin blockade on disease severity in a murine model of limb-girdle muscular dystrophy. Am. J. Pathol. 2006, 168, 1975–1985. [Google Scholar] [CrossRef]
- Braga, M.; Pervin, S.; Norris, K.; Bhasin, S.; Singh, R. Inhibition of In Vitro and In Vivo Brown Fat Differentiation Program by Myostatin. Obesity 2013, 21, 1180–1188. [Google Scholar] [CrossRef]
- Li, X.; Xie, S.S.; Qian, L.L.; Cai, C.B.; Bi, H.F.; Cui, W.T. Identification of genes related to skeletal muscle growth and development by integrated analysis of transcriptome and proteome in myostatin-edited Meishan pigs. J. Proteom. 2020, 213, 103628. [Google Scholar] [CrossRef]
- Xu, K.; Han, C.X.; Zhou, H.; Ding, J.M.; Xu, Z.; Yang, L.Y.; He, C.; Akinyemi, F.; Zheng, Y.M.; Qin, C.; et al. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. Int. J. Mol. Sci. 2020, 21, 2584. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Hansen, R.H.; Hansen, J.B.; Olsen, C.H.; Galle, P.; Mandrup-Poulsen, T.; Gehl, J.; Pedersen, B.K.; Hojman, P. Over-expression of Follistatin-like 3 attenuates fat accumulation and improves insulin sensitivity in mice. Metabolism 2015, 64, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, Y.; Hagiwara, H.; Nakatani, M.; Yasue, A.; Moriyama, K.; Murakami, T.; Tsuchida, K.; Noji, S.; Sunada, Y. Muscular atrophy of caveolin-3-deficient mice is rescued by myostatin inhibition. J. Clin. Invest. 2006, 116, 2924–2934. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Jin, J.P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene 2016, 582, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cancedda, F.D.; Dozin, B.; Zerega, B.; Cermelli, S.; Cancedda, R. Extracellular fatty acid binding protein (ex-FABP) is a stress protein expressed during chondrocyte and myoblast differentiation. Osteoarthr. Cartil. 2001, 9, S118–S122. [Google Scholar]
- Gentili, C.; Cermelli, S.; Tacchetti, C.; Cossu, G.; Cancedda, R.; Cancedda, F.D. Expression of the extracellular fatty acid binding protein (Ex-FABP) during muscle fiber formation in vivo and in vitro. Exp. Cell Res. 1998, 242, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.W.; Hudson, N.; Seo, D.; Lee, S.; Khatri, B.; Lassiter, K.; Cook, D.; Piekarski, A.; Dridi, S.; Anthony, N.; et al. RNA sequencing for global gene expression associated with muscle growth in a single male modern broiler line compared to a foundational Barred Plymouth Rock chicken line. BMC Genom. 2017, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.V.N.; Lamont, S.J.; Rothschild, M.F.; Persia, M.E.; Ashwell, C.M.; Schmidt, C.J. Transcriptome Analysis of Post-Hatch Breast Muscle in Legacy and Modern Broiler Chickens Reveals Enrichment of Several Regulators of Myogenic Growth. PLoS ONE 2015, 10, e0122525. [Google Scholar]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 MAPK signaling pathway: A major regulator of skeletal muscle development. Mol. Cell Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef]
- Chelh, I.; Meunier, B.; Picard, B.; Reecy, M.J.; Chevalier, C.; Hocquette, J.F.; Cassar-Malek, I. Molecular profiles of Quadriceps muscle in myostatin-null mice reveal PI3K and apoptotic pathways as myostatin targets. BMC Genom. 2009, 10, 196. [Google Scholar] [CrossRef]
- Wang, L.M.; Huang, Y.; Wang, X.L.; Chen, Y.L. Label-Free LC-MS/MS Proteomics Analyses Reveal Proteomic Changes Accompanying MSTN KO in C2C12 Cells. Biomed. Res. Int. 2019, 2019, 7052456. [Google Scholar] [CrossRef] [PubMed]
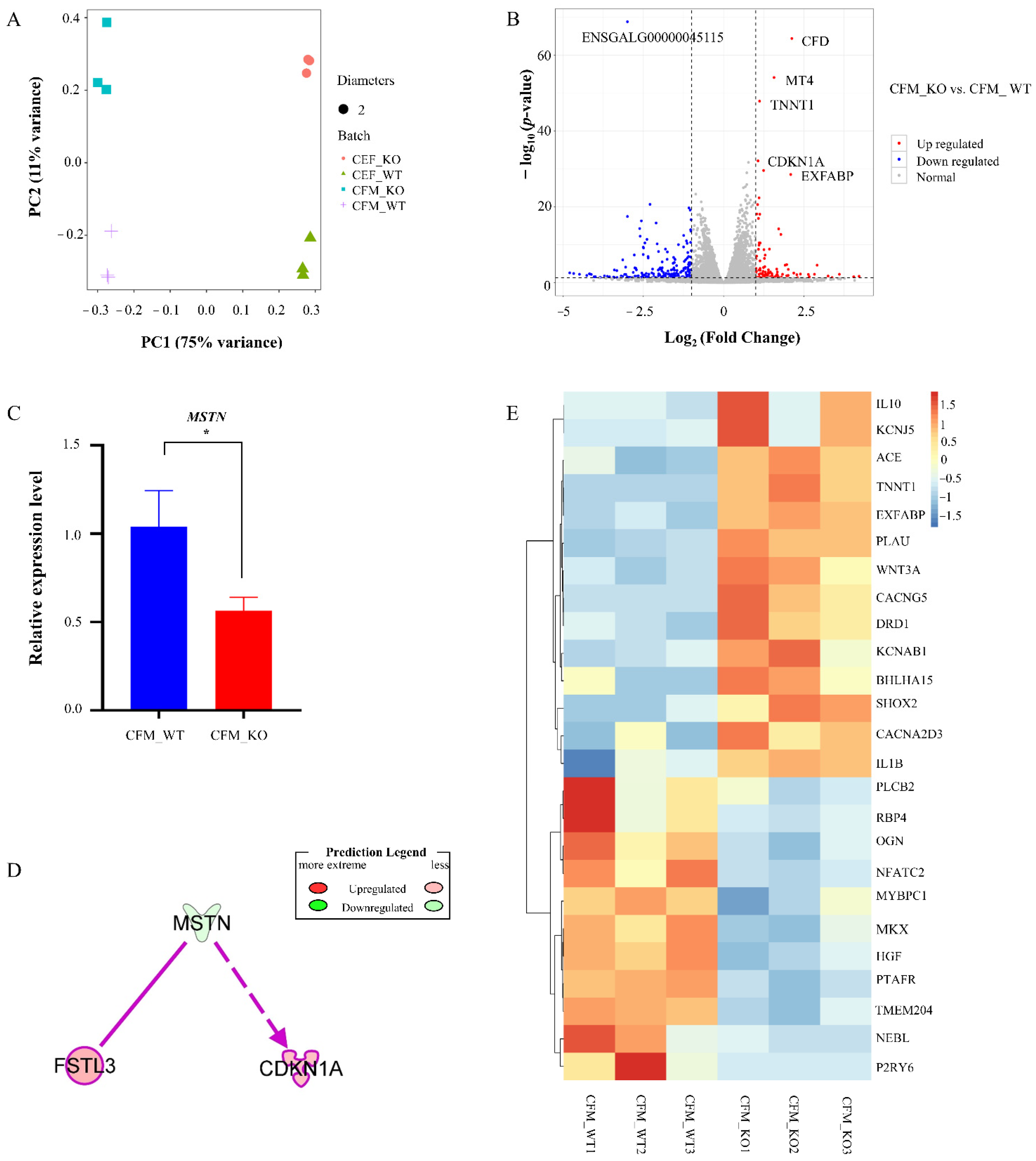
| Gene Stable ID | Gene Name | Log2 Fold Change | p Value |
|---|---|---|---|
| ENSGALG00000045115 | −2.998309986 | 1.38 × 10−69 | |
| ENSGALG00000040832 | CFD | 2.120508849 | 3.47 × 10−65 |
| ENSGALG00000028451 | MT4 | 1.569523444 | 6.71 × 10−55 |
| ENSGALG00000029203 | TNNT1 | 1.116180129 | 1.18 × 10−48 |
| ENSGALG00000028318 | CDKN1A | 1.070316829 | 7.08 × 10−33 |
| ENSGALG00000010741 | MAP1LC3C | 1.241659829 | 2.31 × 10−30 |
| ENSGALG00000043064 | EXFABP | 2.087908822 | 2.64 × 10−29 |
| ENSGALG00000006443 | MADPRT1 | 1.103319425 | 3.97 × 10−23 |
| ENSGALG00000044799 | VSIG4 | −2.295106641 | 2.13 × 10−21 |
| ENSGALG00000053846 | 1.055098394 | 2.42 × 10−21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Zhou, H.; Han, C.; Xu, Z.; Ding, J.; Zhu, J.; Qin, C.; Luo, H.; Chen, K.; Jiang, S.; et al. Transcriptomic Analysis of MSTN Knockout in the Early Differentiation of Chicken Fetal Myoblasts. Genes 2022, 13, 58. https://doi.org/10.3390/genes13010058
Xu K, Zhou H, Han C, Xu Z, Ding J, Zhu J, Qin C, Luo H, Chen K, Jiang S, et al. Transcriptomic Analysis of MSTN Knockout in the Early Differentiation of Chicken Fetal Myoblasts. Genes. 2022; 13(1):58. https://doi.org/10.3390/genes13010058
Chicago/Turabian StyleXu, Ke, Hao Zhou, Chengxiao Han, Zhong Xu, Jinmei Ding, Jianshen Zhu, Chao Qin, Huaixi Luo, Kangchun Chen, Shengyao Jiang, and et al. 2022. "Transcriptomic Analysis of MSTN Knockout in the Early Differentiation of Chicken Fetal Myoblasts" Genes 13, no. 1: 58. https://doi.org/10.3390/genes13010058
APA StyleXu, K., Zhou, H., Han, C., Xu, Z., Ding, J., Zhu, J., Qin, C., Luo, H., Chen, K., Jiang, S., Liu, J., Zhu, W., & Meng, H. (2022). Transcriptomic Analysis of MSTN Knockout in the Early Differentiation of Chicken Fetal Myoblasts. Genes, 13(1), 58. https://doi.org/10.3390/genes13010058







