Molecular Responses to Thermal and Osmotic Stress in Arctic Intertidal Mussels (Mytilus edulis): The Limits of Resilience
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. Salinity and Temperature Experiments
2.3. RNA Extraction and Sequencing
2.4. Bioinformatics
3. Results
3.1. Transcriptome Statistics
3.2. Differential Expression and GO Enrichment
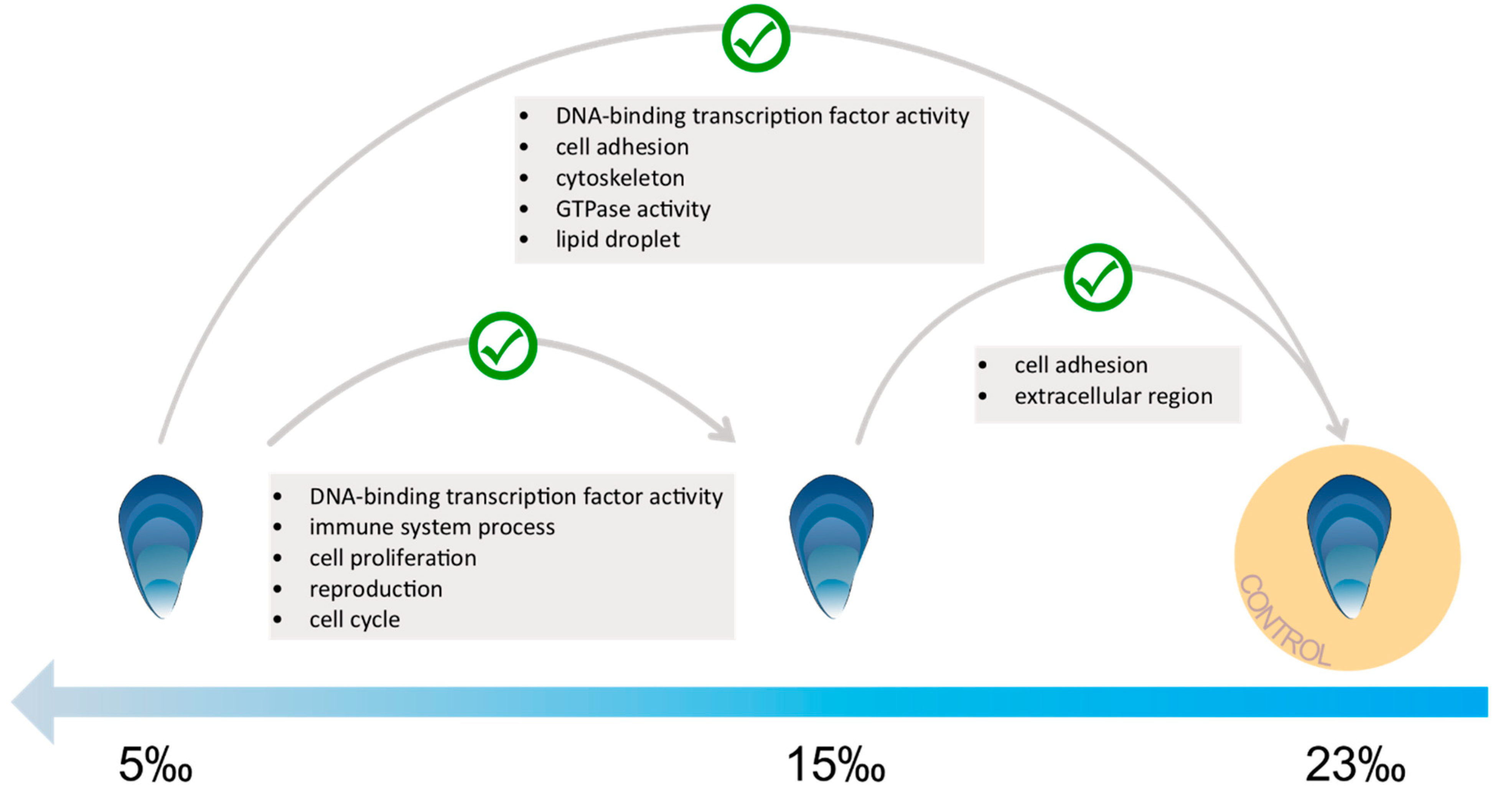
3.3. Low Salinity Exposure
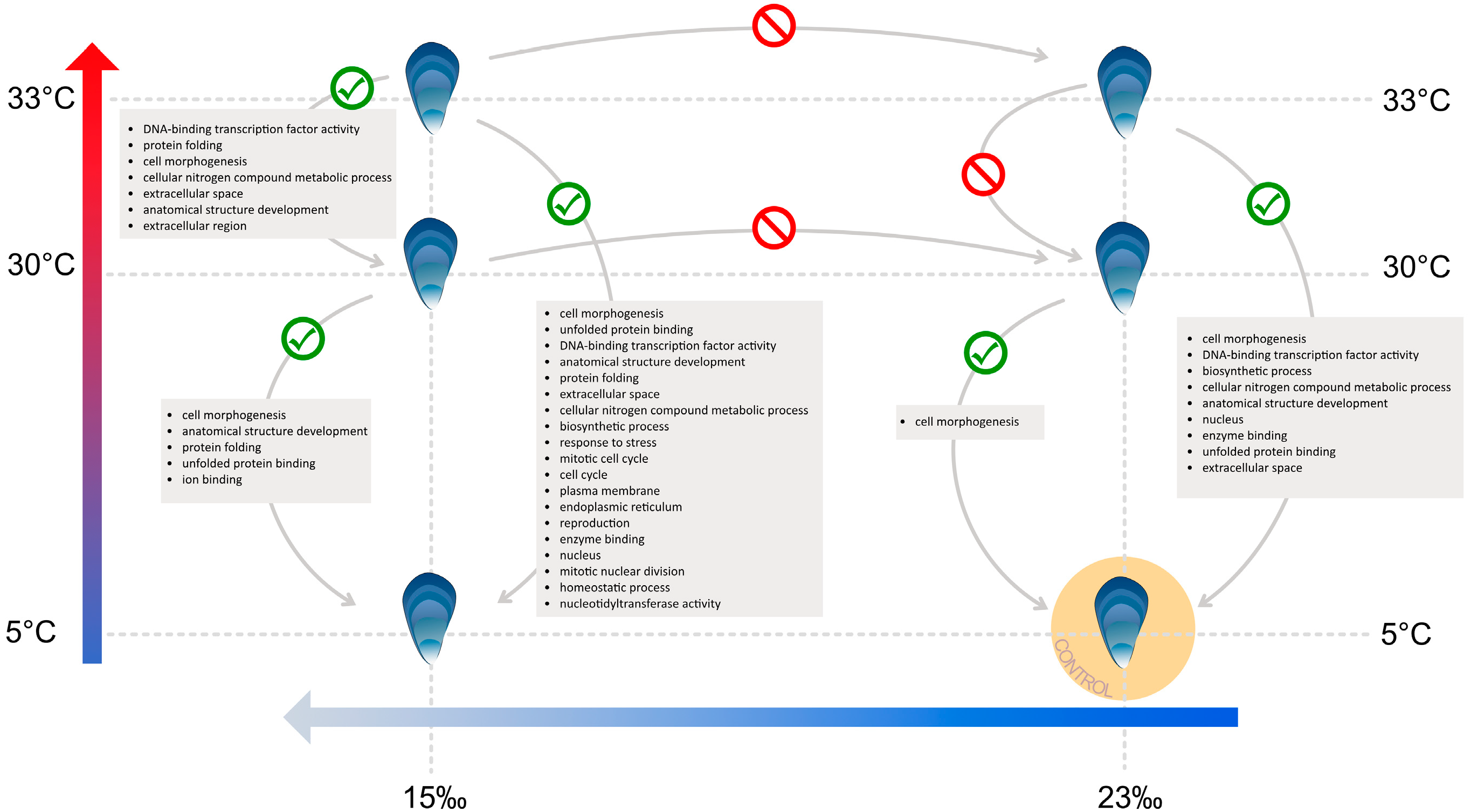
3.4. Heat Stress and Low Salinity Exposure
3.5. Stress-Related Genes
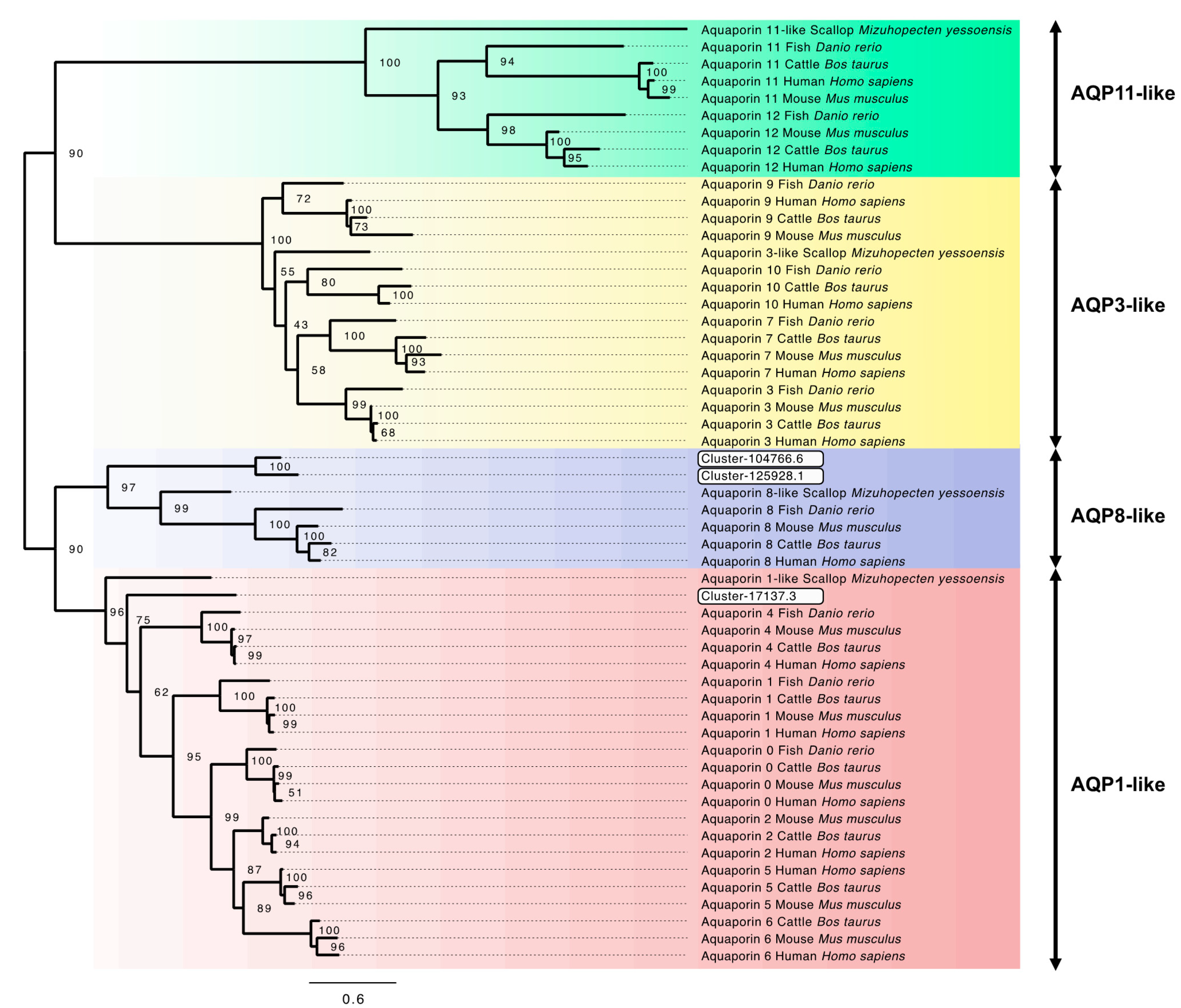
3.6. Aquaporin Genes and Phylogenetic Analysis
4. Discussion
4.1. HSPA12 Genes
4.2. Molecular Response to Low Salinity Exposure
4.3. Cytoskeleton
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clarke, A.; Johnston, N.M. Scaling of metabolic rate with body mass and temperature in teleost fish. J. Anim. Ecol. 1999, 68, 893–905. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Brown, J.H.; West, G.B.; Savage, V.M.; Charnov, E.L. Effects of size and temperature on metabolic rate. Science 2001, 293, 2248–2251. [Google Scholar] [CrossRef] [PubMed]
- Peck, L.S.; Clark, M.S.; Morley, S.A.; Massey, A.; Rossetti, H. Animal temperature limits and ecological relevance: Effects of size, activity and rates of change. Funct. Ecol. 2009, 23, 248–256. [Google Scholar] [CrossRef]
- Peck, L.S. A Cold limit to adaptation in the sea. Trends Ecol. Evol. 2016, 31, 13–26. [Google Scholar] [CrossRef]
- AMAP. Arctic Climate Change Update 2021: Key Trends and Impacts. Summary for Policy-Makers; Arctic Monitoring and Assessment Programme (AMAP): Tromsø, Norway, 2021; 16p. [Google Scholar]
- Kjeldsen, K.K.; Korsgaard, N.J.; Bjørk, A.A.; Khan, S.A.; Box, J.E.; Funder, S.; Larsen, N.K.; Bamber, J.L.; Colgan, W.; van den Broeke, M.; et al. Spatial and temporal distribution of mass loss from the Greenland Ice Sheet since AD 1900. Nature 2015, 528, 396–400. [Google Scholar] [CrossRef]
- Mouginot, J.; Rignot, E.; Bjørk, A.A.; van den Broeke, M.; Millan, R.; Morlighem, M.; Noël, B.; Scheuchl, B.; Wood, M. Forty-six years of Greenland Ice Sheet mass balance from 1972 to 2018. Proc. Natl. Acad. Sci. USA 2019, 116, 9239–9244. [Google Scholar] [CrossRef] [PubMed]
- Sejr, M.K.; Stedmon, C.A.; Bendtsen, J.; Abermann, J.; Juul-Pedersen, T.; Mortensen, J.; Rysgaard, S. Evidence of local and regional freshening of Northeast Greenland coastal waters. Sci. Rep. 2017, 7, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Frölicher, T.L.; Fischer, E.M.; Gruber, N. Marine heatwaves under global warming. Nature 2018, 560, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.; Qiao, F.; Song, Z.; Zhao, J.; Li, X. Projected Freshening of the Arctic Ocean in the 21st Century. J. Geophys. Res. Ocean. 2018, 123, 9232–9244. [Google Scholar] [CrossRef]
- Thyrring, J.; Wegeberg, S.; Blicher, M.E.; Krause-Jensen, D.; Høgslund, S.; Olesen, B.; Jozef, W., Jr.; Mouritsen, K.N.; Peck, L.S.; Sejr, M.K. Latitudinal patterns in intertidal ecosystem structure in West Greenland suggest resilience to climate change. Ecography 2021, 44, 1156–1168. [Google Scholar] [CrossRef]
- Vallière, D.; Guderley, H.; Larochelle, J. Cryoprotective mechanisms in subtidally cultivated and intertidal blue mussels (Mytilus edulis) from the Magdalen Islands, Québec. J. Therm. Biol. 1990, 15, 233–238. [Google Scholar] [CrossRef]
- Williams, R.J. Freezing tolerance in Mytilus edulis. Comp. Biochem. Physiol. 1970, 35, 145–161. [Google Scholar] [CrossRef]
- Thyrring, J.; Tremblay, R.; Sejr, M.K. Local cold adaption increases the thermal window of temperate mussels in the Arctic. Conserv. Physiol. 2019, 7, coz098. [Google Scholar] [CrossRef]
- Thyrring, J.; Blicher, M.E.; Sørensen, J.G.; Wegeberg, S.; Sejr, M.K. Rising air temperatures will increase intertidal mussel abundance in the Arctic. Mar. Ecol. Prog. Ser. 2017, 584, 91–104. [Google Scholar] [CrossRef]
- Nielsen, M.B.; Vogensen, T.K.; Thyrring, J.; Sørensen, J.G.; Sejr, M.K. Freshening increases the susceptibility to heat stress in intertidal mussels (Mytilus edulis) from the Arctic. J. Anim. Ecol. 2021, 90, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.S.; Peck, L.S.; Thyrring, J. Resilience in Greenland intertidal Mytilus: The hidden stress defense. Sci. Total Environ. 2021, 767, 144366. [Google Scholar] [CrossRef]
- Jones, S.J.; Lima, F.P.; Wethey, D.S. Rising environmental temperatures and biogeography: Poleward range contraction of the blue mussel, Mytilus edulis L., in the western Atlantic. J. Biogeogr. 2010, 37, 2243–2259. [Google Scholar] [CrossRef]
- Bayne, B.L. Marine Mussels, Their Ecology and Physiology; Bayne, B.L., Ed.; Cambridge University Press: Cambridge, UK, 1976. [Google Scholar]
- Thyrring, J.; Rysgaard, S.; Blicher, M.E.; Sejr, M.K. Metabolic cold adaptation and aerobic performance of blue mussels (Mytilus edulis) along a temperature gradient into the High Arctic region. Mar. Biol. 2015, 162, 235–243. [Google Scholar] [CrossRef]
- Connor, K.; Gracey, A.Y. Cycles of heat and aerial-exposure induce changes in the transcriptome related to cell regulation and metabolism in Mytilus californianus. Mar. Biol. 2020, 167, 1–12. [Google Scholar] [CrossRef]
- Krogh, A. Osmotic Regulation in Aquatic Animals; Cambridge University Press: Cambridge, UK, 1939. [Google Scholar]
- Davenport, J. Is Mytilus edulis a short term osmoregulator? Comp. Biochem. Physiol. Part. A Physiol. 1979, 64, 91–95. [Google Scholar] [CrossRef]
- Vuorinen, I.; Antsulevich, A.E.; Maximovich, N.V. Spatial distribution and growth of the common mussel Mytilus edulis L. in the archipelago of SW-Finland, northern Baltic Sea. Boreal Environ. Res. 2002, 7, 41–52. [Google Scholar]
- Cawthorne, D.F. A comparative study of the closure responses of some cirripede species exposed to falling seawater concentrations. J. Mar. Biol. Assoc. UK 1979, 59, 811–817. [Google Scholar] [CrossRef]
- Davenport, J. The isolation response of mussels (Mytilus edulis L.) exposed to falling sea-water concentrations. J. Mar. Biol. Assoc. UK 1979, 59, 123–132. [Google Scholar] [CrossRef]
- Duarte, C.M.; Rodriguez-Navarro, A.B.; Delgado-Huertas, A.; Krause-Jensen, D. Dense Mytilus beds along freshwater-influenced greenland shores: Resistance to corrosive waters under high food supply. Estuaries Coasts 2020, 43, 387–395. [Google Scholar] [CrossRef]
- Davenport, J. The opening response of mussels (Mytilus edulis) exposed to rising sea-water concentrations. J. Mar. Biol. Assoc. UK 1981, 61, 667–678. [Google Scholar] [CrossRef]
- Pierce, S.K., Jr.; Greenberg, M.J. The nature of cellular volume regulation in marine bivalves. J. Exp. Biol. 1972, 57, 681–692. [Google Scholar] [CrossRef]
- Deaton, L. Osmotic and ionic regulation in molluscs. In Osmotic and Ionic Regulation: Cells and Animals, 1st ed.; Evans, D.H., Ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2009; pp. 107–134. [Google Scholar]
- Hawkins, A.J.; Hilbish, T.J. The costs of cell volume regulation: Protein metabolism during hyperosmotic adjustment. J. Mar. Biol. Assoc. UK 1992, 72, 569–578. [Google Scholar] [CrossRef]
- Khlebovich, V.V. Acclimation of animal organisms: Basic theory and applied aspects. Biol. Bull. Rev. 2017, 7, 279–286. [Google Scholar] [CrossRef]
- Stickle, W.B.; Sabourin, T.D. Effects of salinity on the respiration and heart rate of the common mussel, Mytilus edulis L., and the black chiton, Katherina tunicata (Wood). J. Exp. Mar. Bio. Ecol. 1979, 41, 257–268. [Google Scholar] [CrossRef]
- Tedengren, M.; Kautsky, N. Comparative study of the physiology and its probable effect on size in blue mussels (Mytilus edulis L.) from the North Sea and the Northern Baltic Proper. Ophelia 1986, 25, 147–155. [Google Scholar] [CrossRef]
- Kautsky, N. Growth and size structure in a Baltic Mytilus edulis L. population. Mar. Biol. 1982, 68, 117–133. [Google Scholar] [CrossRef]
- Telesca, L.; Peck, L.S.; Sanders, T.; Thyrring, J.; Sejr, M.K.; Harper, E.M. Biomineralization plasticity and environmental heterogeneity predict geographical resilience patterns of foundation species to future change. Glob. Chang. Biol. 2019, 25, 4179–4193. [Google Scholar] [CrossRef]
- Almada-Villela, P.C. The effects of reduced salinity on the shell growth of small Mytilus edulis. J. Mar. Biol. Assoc. UK 1984, 64, 171–182. [Google Scholar] [CrossRef]
- Landes, A.; Dolmer, P.; Poulsen, L.K.; Petersen, J.K.; Vismann, B. Growth and respiration in blue mussels (Mytilus spp.) from different salinity regimes. J. Shellfish Res. 2015, 34, 373–382. [Google Scholar] [CrossRef]
- Kinne, O. The effects of temperature and salinity on marine and brackish water animals. II. Salinity and temperature-salinity combinations. Oceanogr. Mar. Biol. Ann. Rev. 1964, 2, 281–339. [Google Scholar]
- Sokolova, I.M. Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr. Comp. Biol. 2013, 53, 597–608. [Google Scholar] [CrossRef]
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Buckley, B.A.; Owen, M.-E.; Hofmann, G.E. Adjusting the thermostat: The threshold induction temperature for the heat-shock response in intertidal mussels (genus Mytilus) changes as a function of thermal history. J. Exp. Biol. 2001, 204, 3571–3579. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, B.L.; Sanders, J.G.; Somero, G.N. Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): Molecular correlates of invasive success. J. Exp. Biol. 2010, 213, 3548–3558. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, B.L.; Somero, G.N. Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol. Ecol. 2011, 20, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhu, Q.; Zhang, L.; Li, C.; Li, L.; She, Z.; Huang, B.; Zhang, G. Genome and transcriptome analyses provide insight into the euryhaline adaptation mechanism of Crassostrea gigas. PLoS ONE 2013, 8, e58563. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36 (Suppl. 1), D480–D484. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35 (Suppl. 2), W182–W185. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- R Studio Team. R Studio: Integrated Development for R; R Studio: Boston, MA, USA, 2016. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 17 December 2021).
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, M.S.; McNutt, P.M. Cadherins and their connections: Adhesion junctions have broader functions. Curr. Opin. Cell Biol. 1999, 11, 554–560. [Google Scholar] [CrossRef]
- McDougall, C.; Hammond, M.J.; Dailey, S.C.; Somorjai, I.M.L.; Cummins, S.F.; Degnan, B.M. The evolution of ependymin-related proteins. BMC Evol. Biol. 2018, 18, 182. [Google Scholar] [CrossRef]
- Pimm, M.L.; Hotaling, J.; Henty-Ridilla, J.L. Chapter Five—Profilin choreographs actin and microtubules in cells and cancer. In Actin Cytoskeleton in Cancer Progression and Metastasis—Part A; Thomas, C., Galluzzi, L.B.T., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 355, pp. 155–204. [Google Scholar] [CrossRef]
- Hamer, B.; Jakšić, Ž.; Pavičić-Hamer, D.; Perić, L.; Medaković, D.; Ivanković, D.; Pavičić, J.; Zilberberg, C.; Schröder, H.C.; Müller, W.E.G.; et al. Effect of hypoosmotic stress by low salinity acclimation of Mediterranean mussels Mytilus galloprovincialis on biological parameters used for pollution assessment. Aquat. Toxicol. 2008, 89, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Gaitanaki, C.; Kefaloyianni, E.; Marmari, A.; Beis, I. Various stressors rapidly activate the p38-MAPK signaling pathway in Mytilus galloprovincialis (Lam.). Mol. Cell. Biochem. 2004, 260, 119–127. [Google Scholar] [CrossRef]
- Kosicka, E.; Lesicki, A.; Pieńkowska, J.R. Molluscan aquaporins: An overview, with some notes on their role in the entry into aestivation in gastropods. Molluscan Res. 2020, 40, 101–111. [Google Scholar] [CrossRef]
- Calcino, A.D.; De Oliveira, A.L.; Simakov, O.; Schwaha, T.; Zieger, E.; Wollesen, T.; Wanninger, A. The quagga mussel genome and the evolution of freshwater tolerance. DNA Res. 2019, 26, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Sejr, M.K.; Mouritsen, K.N.; Krause-Jensen, D.; Olesen, B.; Blicher, M.E.; Thyrring, J. Small scale factors modify impacts of temperature, ice scour and waves and drive rocky intertidal community structure in a Greenland fjord. Front. Mar. Sci. 2021, 7, 1178. [Google Scholar] [CrossRef]
- Kroeker, K.J.; Kordas, R.L.; Harley, C.D.G. Embracing interactions in ocean acidification research: Confronting multiple stressor scenarios and context dependence. Biol. Lett. 2017, 13, 20160802. [Google Scholar] [CrossRef]
- Hazel, J.R.; Eugene Williams, E. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 1990, 29, 167–227. [Google Scholar] [CrossRef]
- Thyrring, J.; Tremblay, R.; Sejr, M.K. Importance of ice algae and pelagic phytoplankton as food sources revealed by fatty acid trophic markers in a keystone species (Mytilus trossulus) from the High Arctic. Mar. Ecol. Prog. Ser. 2017, 572, 155–164. [Google Scholar] [CrossRef][Green Version]
- Evans, T.G.; Kültz, D. The cellular stress response in fish exposed to salinity fluctuations. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Kültz, D. Defining biological stress and stress responses based on principles of physics. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 350–358. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Brunsting, J.F.; Stege, G.J.J.; Burgman, P.W.J.J.; Konings, A.W.T. Thermal protein denaturation and protein aggregation in cells made thermotolerant by various chemicals: Role of heat shock proteins. Exp. Cell Res. 1995, 219, 536–546. [Google Scholar] [CrossRef]
- Todgham, A.E.; Schulte, P.M.; Iwama, G.K. Cross-tolerance in the tidepool sculpin: The role of heat shock proteins. Physiol. Biochem. Zool. 2005, 78, 133–144. [Google Scholar] [CrossRef]
- Collins, M.; Clark, M.S.; Spicer, J.I.; Truebano, M. Transcriptional frontloading contributes to cross-tolerance between stressors. Evol. Appl. 2021, 14, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Truong, Q.A.; Park, S.; Breslow, J.L. Two Hsp70 family members expressed in atherosclerotic lesions. Proc. Natl. Acad. Sci. USA 2003, 100, 1256–1261. [Google Scholar] [CrossRef]
- Brocchieri, L.; Conway de Macario, E.; Macario, A.J.L. Hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef]
- Cheng, J.; Xun, X.; Kong, Y.; Wang, S.; Yang, Z.; Li, Y.; Kong, D.; Wang, S.; Zhang, L.; Hu, X.; et al. Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish. Shellfish Immunol. 2016, 58, 266–273. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef]
- Davenport, J. Osmotic control in marine animals. Symp. Soc. Exp. Biol. 1985, 39, 207–244. [Google Scholar]
- Hauton, C. Effects of salinity as a stressor to aquatic invertebrates. In Stressors in the Marine Environment; Solan, M., Whiteley, N., Eds.; Oxford University Press: Oxford, UK, 2016. [Google Scholar] [CrossRef]
- Kirschner, L.B. Water and ions. In Environmental and Metabolic Animal Physiology; Prosser, C.L., Ed.; Wiley-Liss: New York, NY, USA, 1991; pp. 13–107. [Google Scholar]
- Lovett, D.L.; Verzi, M.P.; Burgents, J.E.; Tanner, C.A.; Glomski, K.; Lee, J.J.; Towle, D.W. Expression profiles of Na+,K+-ATPase during acute and chronic hypo-osmotic stress in the blue crab Callinectes sapidus. Biol. Bull. 2006, 211, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Marom, S. Slow changes in the availability of voltage-gated ion channels: Effects on the dynamics of excitable membranes. J. Membr. Biol. 1998, 161, 105–113. [Google Scholar] [CrossRef]
- Finn, R.N.; Cerdà, J. Evolution and functional diversity of aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Agre, P.; Preston, G.M.; Smith, B.L.; Jung, J.S.; Raina, S.; Moon, C.; Guggino, W.B.; Nielsen, S. Aquaporin CHIP: The archetypal molecular water channel. Am. J. Physiol. Physiol. 1993, 265, F463–F476. [Google Scholar] [CrossRef]
- Tani, T.; Tominaga, T.; Allen, R.D.; Naitoh, Y. Development of periodic tension in the contractile vacuole complex membrane of Paramecium governs its membrane dynamics. Cell Biol. Int. 2002, 26, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Huebert, R.C.; Splinter, P.L.; Garcia, F.; Marinelli, R.A.; Larusso, N.F. Expression and localization of aquaporin water channels in rat hepatocytes. Evidence for a role in canalicular bile secretion. J. Biol. Chem. 2002, 277, 22710–22717. [Google Scholar] [CrossRef]
- Pedersen, S.F.; Hoffmann, E.K.; Mills, J.W. The cytoskeleton and cell volume regulation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001, 130, 385–399. [Google Scholar] [CrossRef]
- Tang, R.; Lan, P.; Ding, C.; Wang, J.; Zhang, T.; Wang, X. A new perspective on the toxicity of arsenic-contaminated soil: Tandem mass tag proteomics and metabolomics in earthworms. J. Hazard. Mater. 2020, 398, 122825. [Google Scholar] [CrossRef]
- Xu, L.; Peng, X.; Yu, D.; Ji, C.; Zhao, J.; Wu, H. Proteomic responses reveal the differential effects induced by cadmium in mussels Mytilus galloprovincialis at early life stages. Fish. Shellfish Immunol. 2016, 55, 510–515. [Google Scholar] [CrossRef]
- Ji, C.; Wu, H.; Wei, L.; Zhao, J. ITRAQ-based quantitative proteomic analyses on the gender-specific responses in mussel Mytilus galloprovincialis to tetrabromobisphenol A. Aquat. Toxicol. 2014, 157, 30–40. [Google Scholar] [CrossRef]
- Leicht, B.G.; Biessmann, H.; Palter, K.B.; Bonner, J.J. Small heat shock proteins of Drosophila associate with the cytoskeleton. Proc. Natl. Acad. Sci. USA 1986, 83, 90–94. [Google Scholar] [CrossRef]
- Dasgupta, S.; Hohman, T.C.; Carper, D. Hypertonic stress induces αB-crystallin expression. Exp. Eye Res. 1992, 54, 461–470. [Google Scholar] [CrossRef]
- Neuhofer, W.; Müller, E.; Burger-Kentischer, A.; Fraek, M.L.; Thurau, K.; Beck, F.X. Pretreatment with hypertonic NaCl protects MDCK cells against high urea concentrations. Pflug. Arch. Eur. J. Physiol. 1998, 435, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Beck, F.-X.; Neuhofer, W.; Müller, E. Molecular chaperones in the kidney: Distribution, putative roles, and regulation. Am. J. Physiol. Physiol. 2000, 279, F203–F215. [Google Scholar] [CrossRef] [PubMed]

| Assembly Statistics | |
|---|---|
| Number of nucleotides (nt) | 394,372,995 |
| Number of unigenes | 449,638 |
| Unigene minimum length (bp) | 301 |
| Unigene mean length (bp) | 877 |
| Unigene medium length (bp) | 621 |
| Unigene maximum length (bp) | 17,583 |
| N50 | 1104 |
| N90 | 420 |
| Number of unigenes between 200–500 bp | 165,273 |
| Number of unigenes between 500–1k bp | 163,324 |
| Number of unigenes between 1k–2k bp | 89,465 |
| Annotation Rates | |
| Database | % annotation |
| NR (NCBI non-redundant protein sequences) | 35.08 |
| NT (NCBI nucleotide sequences) | 9.75 |
| KEGG (Kyoto Encyclopaedia of Genes and Genomes Orthologues) | 5.4 |
| Swiss-Prot: (Manually annotated protein sequences) | 19.35 |
| Pfam: (Protein domains and families) | 12.33 |
| GO: (Gene Ontology) | 9.54 |
| KOG: (euKaryotic Orthologous Groups) | 7.96 |
| Annotated in all Databases | 0.51 |
| Annotated in at least one Database | 40.68 |
| Comparison | Upregulated Genes | Downregulated Genes | Total Genes |
|---|---|---|---|
| Low salinity exposure | |||
| Low salinity vs. control | 5004 | 3768 | 8772 |
| Very low salinity vs. control | 17,267 | 6824 | 24,091 |
| Very low salinity vs. low salinity | 10,466 | 5354 | 15,820 |
| Heat stress + low salinity exposure | |||
| 30 °C vs. control | 7002 | 4195 | 11,197 |
| 33 °C vs. control | 7033 | 8824 | 15,857 |
| 33 °C vs. 30 °C | 3958 | 8314 | 12,272 |
| Low salinity + 30 °C vs. 30 °C | 3781 | 4734 | 8515 |
| Low salinity + 33 °C vs. 33 °C | 6243 | 2802 | 9045 |
| Low salinity + 33 °C vs. low salinity + 30 °C | 7506 | 4948 | 12,454 |
| Low salinity + 30 °C vs. low salinity | 4722 | 4540 | 9262 |
| Low salinity + 33 °C vs. low salinity | 10,796 | 7202 | 17,998 |
| Gene Type | Comparisons | |||||
|---|---|---|---|---|---|---|
| Low Salinity vs. Control | Very Low Salinity vs. Control | Very Low Salinity vs. Low Salinity | ||||
| ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | |
| Ion channel: Potassium channel | 2 | 2 | 14 | 0 | 5 | 1 |
| Ion channel: Chloride channel | 0 | 0 | 4 | 0 | 2 | 0 |
| Ion channel: Calcium channel | 1 | 0 | 6 | 2 | 4 | 1 |
| Ion exchangers | 1 | 1 | 8 | 0 | 4 | 0 |
| Aquaporins | 0 | 1 | 1 | 0 | 2 | 0 |
| ATPase pumps (V-type) | 0 | 2 | 3 | 2 | 3 | 0 |
| ATPase pumps (Other) | 0 | 0 | 11 | 1 | 3 | 0 |
| Osmolyte transporters (SLC6A5/SLC6A9) | 4 | 0 | 17 | 5 | 7 | 0 |
| Gene Family | Family Member | Comparisons | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 30 °C vs. Control | 33 °C vs. Control | Low Sal. + 30 °C vs. 30 °C | Low Sal. + 33 °C vs. 33 °C | Low Sal. + 30 °C vs. Low Salinity | Low Sal. + 33 °Cvs. Low Salinity | Low Salinity vs. Control | Very Low Salinity vs. Control | Very Low Salinity vs. Low Salinity | ||
| Catalase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Caspase | 16 | 8 | 9 | 13 | 10 | 16 | 14 | 51 | 39 | |
| Glutathione | 12 | 8 | 7 | 13 | 5 | 15 | 4 | 14 | 11 | |
| Hypoxia | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 2 | 1 | |
| Superoxide dismutase | 4 | 8 | 5 | 6 | 4 | 11 | 4 | 5 | 1 | |
| Thioredoxin | 5 | 4 | 3 | 3 | 2 | 7 | 7 | 11 | 1 | |
| p38 mitogen-activated protein kinases | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Heat shock proteins | HSPA12A | 27 | 11 | 15 | 21 | 17 | 21 | 18 | 61 | 54 |
| HSPA12B | 12 | 9 | 20 | 20 | 19 | 22 | 9 | 59 | 48 | |
| small HSP | 24 | 24 | 0 | 1 | 22 | 23 | 1 | 22 | 13 | |
| HSP68 | 23 | 23 | 1 | 1 | 24 | 22 | 0 | 19 | 16 | |
| HSPA5 | 1 | 10 | 0 | 0 | 7 | 12 | 0 | 0 | 0 | |
| HSP70B2 | 8 | 11 | 2 | 1 | 9 | 10 | 0 | 9 | 9 | |
| HSC70 | 2 | 4 | 1 | 0 | 6 | 5 | 1 | 3 | 7 | |
| HSP90 | 0 | 0 | 0 | 1 | 7 | 10 | 1 | 1 | 3 | |
| HSP110 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| HSP70 Family | 39 | 29 | 4 | 4 | 31 | 36 | 1 | 28 | 27 | |
| HSP total | 137 | 121 | 43 | 49 | 143 | 162 | 31 | 202 | 177 | |
| All stress genes | 175 | 149 | 67 | 86 | 164 | 212 | 60 | 285 | 230 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrett, N.J.; Thyrring, J.; Harper, E.M.; Sejr, M.K.; Sørensen, J.G.; Peck, L.S.; Clark, M.S. Molecular Responses to Thermal and Osmotic Stress in Arctic Intertidal Mussels (Mytilus edulis): The Limits of Resilience. Genes 2022, 13, 155. https://doi.org/10.3390/genes13010155
Barrett NJ, Thyrring J, Harper EM, Sejr MK, Sørensen JG, Peck LS, Clark MS. Molecular Responses to Thermal and Osmotic Stress in Arctic Intertidal Mussels (Mytilus edulis): The Limits of Resilience. Genes. 2022; 13(1):155. https://doi.org/10.3390/genes13010155
Chicago/Turabian StyleBarrett, Nicholas J., Jakob Thyrring, Elizabeth M. Harper, Mikael K. Sejr, Jesper G. Sørensen, Lloyd S. Peck, and Melody S. Clark. 2022. "Molecular Responses to Thermal and Osmotic Stress in Arctic Intertidal Mussels (Mytilus edulis): The Limits of Resilience" Genes 13, no. 1: 155. https://doi.org/10.3390/genes13010155
APA StyleBarrett, N. J., Thyrring, J., Harper, E. M., Sejr, M. K., Sørensen, J. G., Peck, L. S., & Clark, M. S. (2022). Molecular Responses to Thermal and Osmotic Stress in Arctic Intertidal Mussels (Mytilus edulis): The Limits of Resilience. Genes, 13(1), 155. https://doi.org/10.3390/genes13010155







