Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Mitogenomes Sequence Analysis
2.3. Phylogenetic Analysis
3. Results
3.1. Mitogenome Organization and Gene Content
3.2. Protein-Coding Genes and Relative Synonymous Codon Usage
3.3. Transfer and Ribosomal RNA Genes
3.4. Control Region
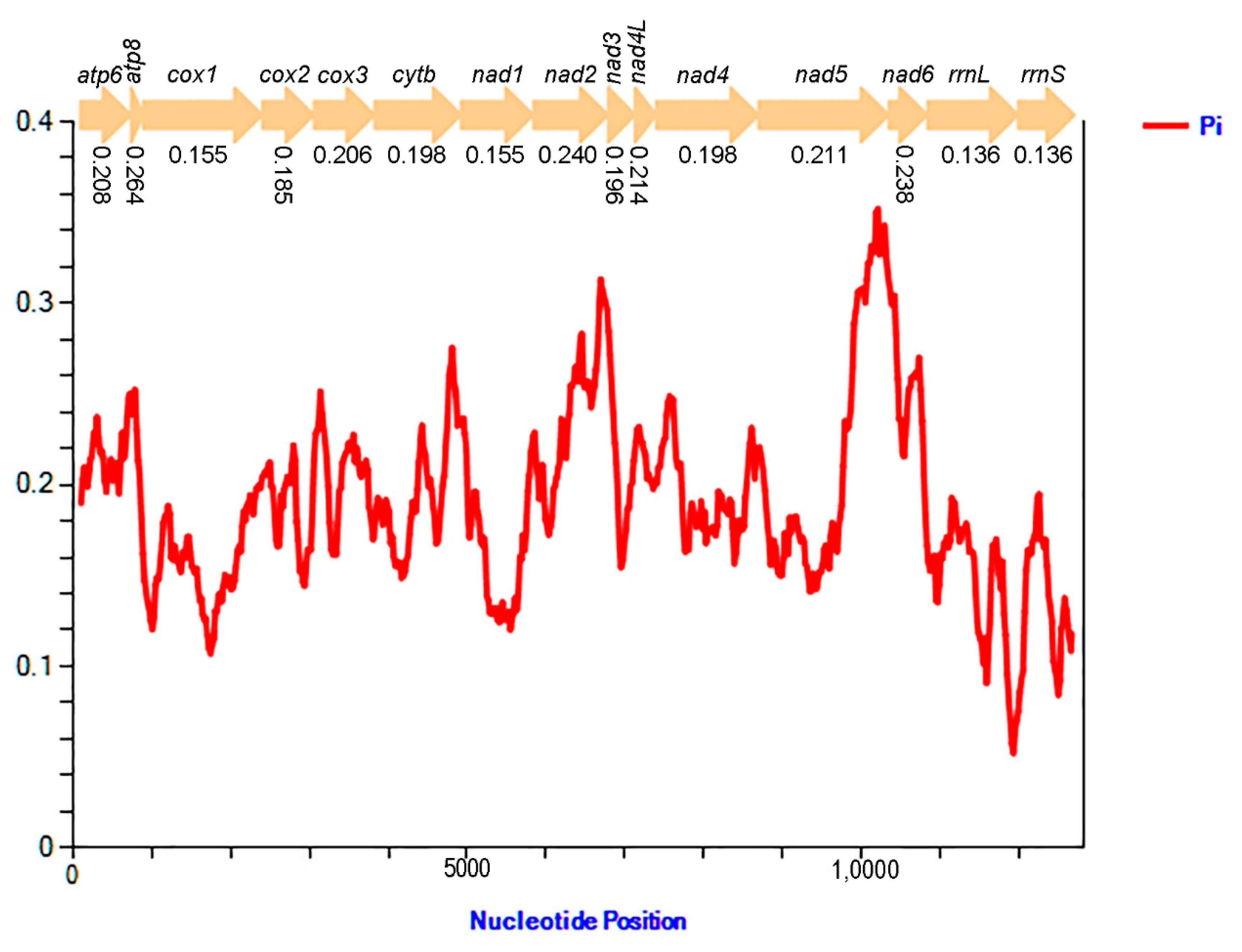
3.5. Nucleotide Diversity
3.6. Phylogenetic Analyses
4. Discussion
4.1. Comparative Analysis of Fulgorid Mitogenomes
4.2. Nucleotide Diversity of Fulgorid Mitogenomtes
4.3. Phylogeny and Species Delimitation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bourgoin, T. FLOW (Fulgoromorpha Lists on the Web), Version 8. Available online: http://hemipteradatabases.org/flow/ (accessed on 6 June 2021).
- Metcalf, Z.P. General Catalogue of the Hemiptera; Fasc. IV: Fulgoroidea. Part. 9: Fulgoridae; North Carolina State College: Raleigh, NC, USA, 1947. [Google Scholar]
- Mason, R.T.; Fales, H.M.; Jones, T.H.; O’Brien, L.B.; Taylor, T.W.; Hogue, C.L.; Blum, M.S. Characterization of fulgorid waxes (Homoptera: Fulgoroidea: Insecta). Insect Biochem. 1989, 19, 737–740. [Google Scholar] [CrossRef]
- Goemans, G. The Fulgoridae (Hemiptera, Fulgoromorpha) of Guatemala. In Biodiversidad de Guatemala; Cano, E.B., Ed.; The Universidad del Valle de Guatemala: Guatemala City, Guatemala, 2006; Volume 1, pp. 337–344. [Google Scholar]
- Lee, D.H.; Park, Y.L.; Leskey, T.C. A review of biology and management of Lycorma delicatula (Hemiptera: Fulgoridae), an emerging global invasive species. J. Asia Pac. Entomol. 2019, 22, 589–596. [Google Scholar] [CrossRef]
- Urban, J.M. Perspective: Shedding light on spotted lanternfly impacts in the USA. Pest Manag. Sci. 2019, 76, 10–17. [Google Scholar] [CrossRef]
- Urban, J.M.; Cryan, J.R. Entomologically famous, evolutionarily unexplored: The first phylogeny of the lanternfly family Fulgoridae (Insecta: Hemiptera: Fulgoroidea). Mol. Phylogenetics Evol. 2009, 50, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Constant, J.; Pham, H.T. Review of the clavatus group of the lanternfly genus Pyrops (Hemiptera: Fulgoromorpha: Fulgoridae). Eur. J. Taxon. 2017, 305, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Kirkaldy, G.W. Bibliographical and nomenclatorial notes on the Hemiptera No. 3. Entomologist 1904, 37, 279–283. [Google Scholar] [CrossRef]
- Muir, F.A.G. On the classification of the Fulgoroidea (Homoptera). In Proceedings of the Hawaiian Entomological Society, Honolulu, HI, USA, 6 April 1922; Volume 5, pp. 205–247. [Google Scholar]
- Metcalf, Z.P. The Fulgorina of Barro Colorado and other parts of Panama. In Bulletin of the Museum of Comparative Zoology at Harvard College; The Museum: Cambridge, MA, USA, 1938; Volume 82, pp. 277–423. [Google Scholar] [CrossRef]
- Emeljanov, A.F. The problem of differentiation of the families Fulgoridae and Dictyopharidae. Tr. Zool. Inst. Akad. Nauk SSSR 1979, 82, 3–22. [Google Scholar]
- Emeljanov, A.F. Improved Tribal Delimitation of the Subfamily Dictyopharinae and Description of New Genera and New Species (Homoptera, Fulgoroidea, Dictyopharidae). Entomol. Rev. 2011, 90, 299–328. [Google Scholar] [CrossRef]
- Emeljanov, A.F. A new species of the genus Pibrocha Kirkaldy (Homoptera, Fulgoridae) with notes on the systematics of the subfamily Dichopterinae and with description of a new tribe. Entomol. Rev. 2013, 92, 123–129. [Google Scholar] [CrossRef]
- Melichar, L. Monographie der Dictyophorinen (Homoptera). Abh. K. K. Zool. Bot. Ges. Wien Wien 1912, 7, 1–221. [Google Scholar]
- Wang, W.Q.; Xu, S.L.; Constant, J.; Qin, D.Z. Revision of the lanternfly genus Limois Stål, 1863 (Hemiptera: Fulgoromorpha: Fulgoridae) with description of a new species from China. Eur. J. Taxon. 2021, 720, 35–61. [Google Scholar] [CrossRef]
- Constant, J.; Pham, H.T. Extension of the lanternfly genus Neoalcathous Wang and Huang, 1989 to Vietnam with a new species and new subfamily placement (Hemiptera: Fulgoromorpha: Fulgoridae). Belg. J. Entomol. 2018, 59, 1–11. [Google Scholar]
- Constant, J. The lanternfly genus Penthicodes: Key to the species and review of the “Ereosoma group” with two new species and one new subspecies (Hemiptera: Fulgoromorpha: Fulgoridae). Zootaxa 2010, 2523, 1–26. [Google Scholar] [CrossRef]
- Nagai, S.; Porion, T. Fulgoridae 2: Catalogue Illustré des Faunes Asiatique et Australienne; Sciences Nat: Compiègne, France, 1996; p. 80. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Havas, S.S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liang, A.P.; Bu, C.P. A Molecular Phylogeny of Hemiptera Inferred from Mitochondrial Genome Sequences. PLoS ONE 2012, 7, e48778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.Q.; Huang, Y.X.; Bartlett, C.R.; Zhou, F.M.; Meng, R.; Qin, D.Z. Characterization of the complete mitochondrial genomes of two species of the genus Aphaena Guérin-Méneville (Hemiptera: Fulgoridae) and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 141, 29–40. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.V.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.; Pfeiffer, W.T.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; Institute of Electrical and Electronics Engineers (IEEE): New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Huang, Y.X.; Qin, D.Z. Sequencing and analysis of the complete mitochondrial genome of Changeondelphax velitchkovskyi (Hemiptera: Fulgoroidea). Mitochondrial DNA Part B 2018, 3, 90–91. [Google Scholar] [CrossRef] [Green Version]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondrial. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Bui, M. IQ-TREE Manual: Frequently Asked Questions (WWW Document). Available online: http://www.iqtree.org/doc/Frequently-Asked-Questions (accessed on 23 March 2021).
- Urban, J.M.; Cryan, J.R. Evolution of the planthoppers (Insecta: Hemiptera: Fulgoroidea). Mol. Phylogenetics Evol. 2007, 42, 556–572. [Google Scholar] [CrossRef]
- Song, N.; Liang, A.P. A preliminary molecular phylogeny of planthoppers (Hemiptera: Fulgoroidea) based on nuclear and mitochondrial DNA sequences. PLoS ONE 2013, 8, e58400. [Google Scholar] [CrossRef] [Green Version]
- Song, N.; Liang, A.P. The complete mitochondrial genome sequence of Geisha distinctissima (Hemiptera: Flatidae) and comparison with other hemipteran insects. Acta Biochim. Biophys. Sin. 2009, 41, 206–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Liang, A.P. Complete mitochondrial genome of the small brown planthopper, Laodelphax striatellus (Delphacidae: Hemiptera), with a novel gene order. Zool. Sci. 2009, 26, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liang, A.P.; Ma, C. The complete mitochondrial genome sequence of the planthopper, Sivaloka damnosus. J. Insect Sci. 2010, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.J.; Zhu, W.C.; Rong, X.; Ding, X.L.; Zhang, Y.K.; Liu, J.; Chen, D.S.; Du, Y.; Hong, X.Y. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: Conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genom. 2013, 14, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.; Peng, X.X.; Jing, S.L.; Liu, B.F.; Zhu, L.L.; He, G.C. Intraspecific and interspecific variations in the mitochondrial genomes of Nilaparvata (Hemiptera: Delphacidae). J. Econ. Entomol. 2015, 108, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.X.; Guan, D.L.; Niu, Y.; Sang, L.Q.; Zhang, X.X.; Xu, S.Q. Characterization of the complete mitochondrial genome of the Asian planthopper Ricania speculum (Hemiptera: Fulgoroidea: Ricannidae). Conserv. Genet. Resour. 2016, 8, 463–466. [Google Scholar] [CrossRef]
- Xu, S.Y.; Long, J.K.; Chen, X.S. Comparative analysis of the complete mitochondrial genomes of five Achilidae species (Hemiptera: Fulgoroidea) and other Fulgoroidea reveals conserved mitochondrial genome organization. PeerJ 2019, 7, e6659. [Google Scholar] [CrossRef]
- Du, Z.Y.; Wu, Y.F.; Chen, Z.; Cao, L.M.; Ishikawa, T.; Kamitani, S.; Sota, T.; Song, F.; Tian, L.; Cai, W.Z.; et al. Global Phylogeography and Invasion History of the Spotted Lanternfly Revealed by Mitochondrial Phylogenomics. Evol. Appl. 2020, 14, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, A.; Nobandegani, F.F. Detection and evaluation of hydrogen bond strength in nucleic acid base pairs. J. Phys. Chem. A 2008, 112, 281–295. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, X.L.; Qiao, G.X. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE 2013, 8, e77511. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, W.L.; Claessens, A.; Otto, T.D.; Kekre, M.; Fairhurst, R.M.; Rayner, J.C.; Kwiatkowski, D. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017, 45, 1889–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Huang, X.; Deng, J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020, 158, 854–864. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Lunt, D.H.; Zhang, D.X.; Szymura, J.M.; Hewltt, O.M. The insect cytochrome oxidase I gene: Evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol. Biol. 1996, 5, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Shiyang, K.; Vaidya, G.; Ng, P.K.L. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Whitworth, T.L.; Dawson, R.D.; Magalon, H.; Baudry, E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc. R. Soc. B Biol. Sci. 2007, 274, 1731–1739. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Huang, J. Notes of new genus and species of Fulgoroidea from China (Homoptera: Fulgoroidea) (2). Acta Agric. Boreali Sin. 1989, S1, 127–132. [Google Scholar]
- Constant, J. Review of the effusus group of the lanternfly genus Pyrops Spinola, 1839, with one new species and notes on trophobiosis (Hemiptera: Fulgoromorpha: Fulgoridae). Eur. J. Taxon. 2015, 128, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Lallemand, V. Revision des Fulgoridae (Homoptera). Deuxième partie. Faunes Asiatique et Australienne. In Mémoires de l’Institut Royal des Sciences Naturelles de Belgique; Deuxième Série; Koninklijk Belgisch instituut voor Natuur Wetenschappen: Brussel, Belgisch, 1963; Volume 75, pp. 1–99. [Google Scholar]
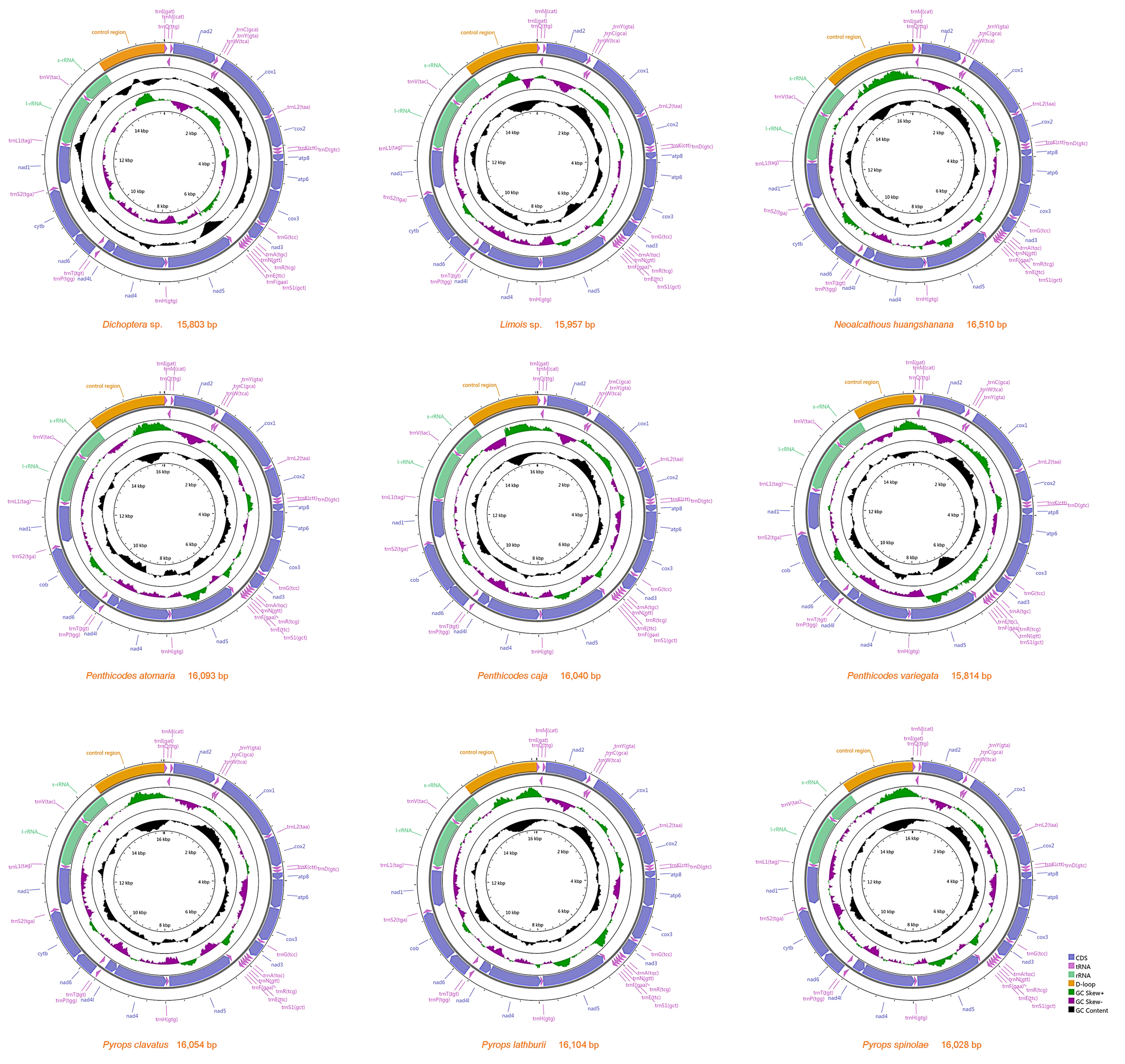
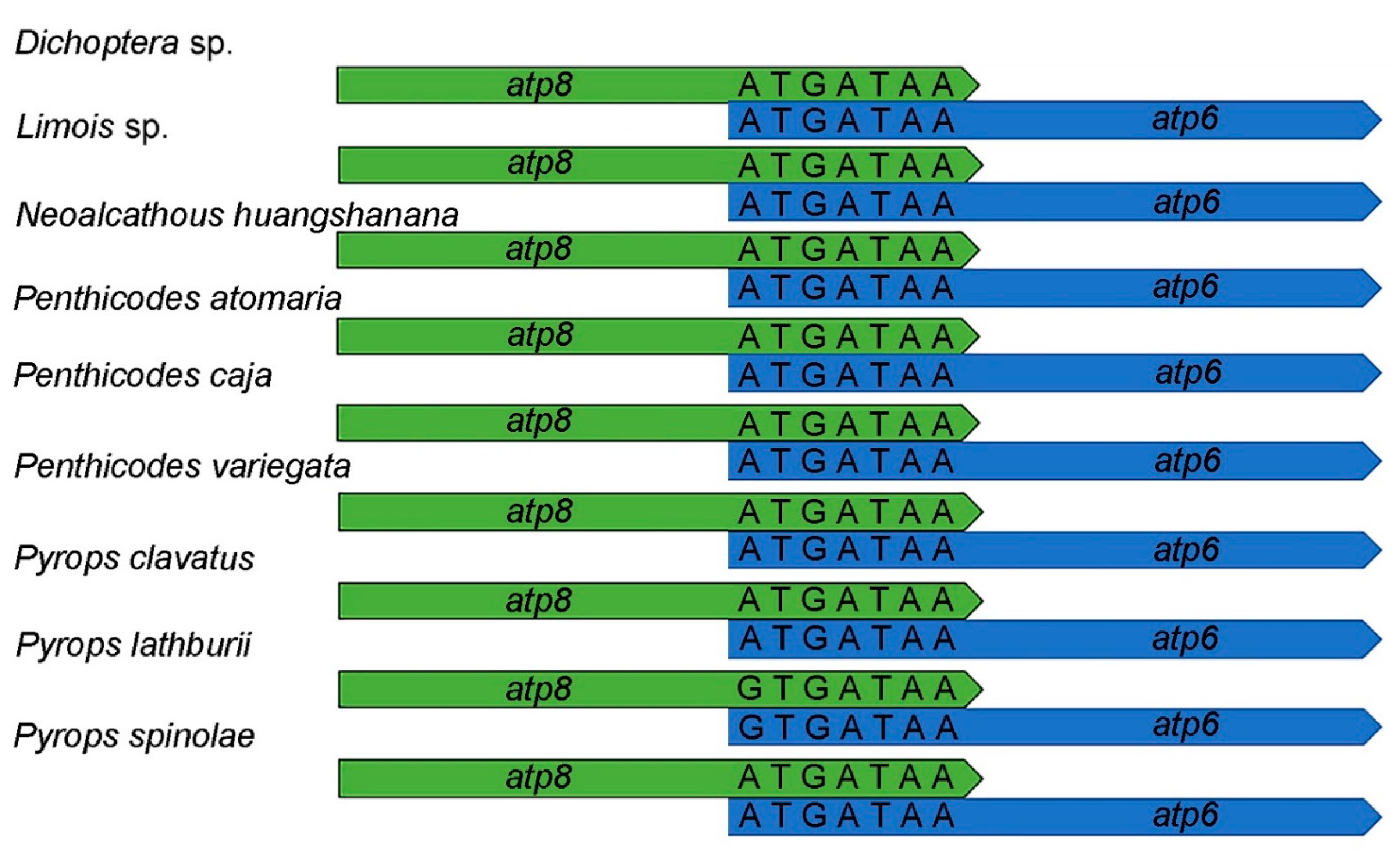

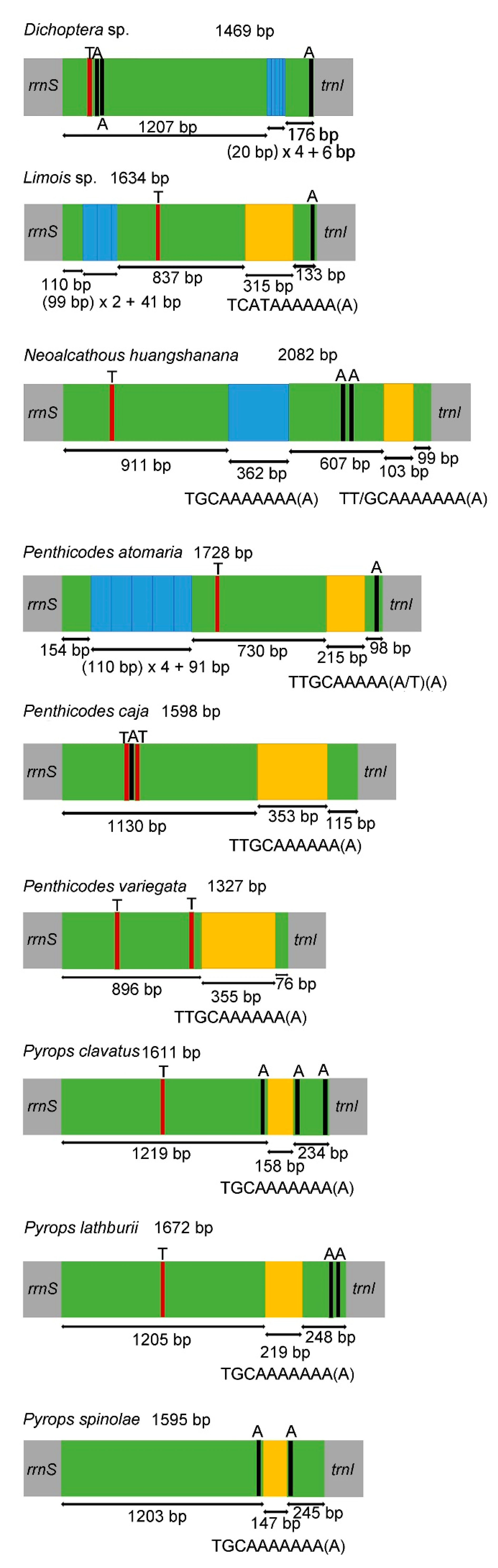

| Superfamily | Family | Subfamily | Species | Accession Number |
|---|---|---|---|---|
| Fulgoroidea | Delphacidae | Delphacinae | Nilaparvata lugens (Stål) | JX880069 |
| Delphacinae | Changeondelphax velitchkovskyi (Melichar) | MG049916 | ||
| Delphacinae | Peregrinus maidis (Ashmead) | MG049917 | ||
| Delphacinae | Nilaparvata bakeri (Muir) | KC333655 | ||
| Delphacinae | Sogatella furcifera (Horváth) | KC512914 | ||
| Delphacinae | Saccharosydne procerus (Matsumura) | MG515237 | ||
| Delphacinae | Bambusiphaga citricolorata Huang and Tian | MH293452 | ||
| Delphacinae | Bambusiphaga furca Huang and Tian | MH293453 | ||
| Delphacinae | Bambusiphaga luodianensis Ding | MH293454 | ||
| Delphacinae | Bambusiphaga maculate Chen and Li | MH293455 | ||
| Delphacinae | Bambusiphaga taibaishana Qin | MH293456 | ||
| Delphacinae | Epeurysa nawaii Matsumura | MH293459 | ||
| Delphacinae | Malaxella flava Ding and Hu | MH293463 | ||
| Delphacinae | Purohita sinica Huang and Ding | MH293467 | ||
| Delphacinae | Tropidocephala brunnipennis Signoret | MH293471 | ||
| Delphacinae | Cemus sauteri (Muir) | MH293457 | ||
| Delphacinae | Chloriona tateyamana Matsumura | MH293458 | ||
| Delphacinae | Ishiharodelphax matsuyamensis (Ishihara) | MH293461 | ||
| Delphacinae | Muirodelphax atratus Vilbaste | MH293464 | ||
| Delphacinae | Numata muiri (Kirkaldy) | MH293465 | ||
| Delphacinae | Perkinsiella saccharicida Kirkaldy | MH293466 | ||
| Delphacinae | Sogata hakonensis (Matsumura) | MH293468 | ||
| Delphacinae | Sogatella vibix (Haupt) | MG515238 | ||
| Delphacinae | Mahmutkashgaria sulcatus (Ding) | MH293470 | ||
| Criomorphinae | Laodelphax striatellus (Fallén) | FJ360695 | ||
| Stenocraninae | Stenocranus matsumurai Metcalf | MH293469 | ||
| Asiracinae | Ugyops sp. | MH352481 | ||
| Lophopidae | Lophops carinata (Kirby) | MT990448 | ||
| Ricaniidae | Ricaniinae | Ricania speculum (Walker) | KX371891 | |
| Ricaniidae | Ricaniinae | Ricania marginalis (Walker) | JN242415 | |
| Ricaniinae | Ricania shantungensis (Chou and Lu) | NC_051496 | ||
| Flatidae | Flatinae | Geisha distinctissima (Walker) | FJ230961 | |
| Issidae | Issinae | Sivaloka damnosus (Chou and Lu) | FJ360694 | |
| Hemisphaeriinae | Hemisphaerius rufovarius Walker | MT210096 | ||
| Achilidae | Achilinae | Betatropis formosana Matsumura | MH324927 | |
| Achilinae | Magadhaideus sp. | MH324928 | ||
| Achilidae sp. | MH324929 | |||
| Plectoderini sp. | MH324930 | |||
| Achilinae | Paracatonidia sp. | MH324931 | ||
| Fulgoridae | Aphaeninae | Lycorma delicatula (White) | EU909203 | |
| Aphaeninae | Lycorma meliae Kato | MT079725 | ||
| Aphaeninae | Aphaena discolor nigrotibiata Schmidt | MN025523 | ||
| Aphaeninae | Aphaena amabilis (Hope) | MN025522 | ||
| Aphaeninae | Penthicodes atomaria (Weber) | MW662662 | ||
| Aphaeninae | Penthicodes variegata (Guérin-Méneville) | MW662664 | ||
| Aphaeninae | Penthicodes caja (Walker) | MW662663 | ||
| Aphaeninae | Limois sp. | MW662660 | ||
| Aphaeninae | Neoalcathous huangshanana Wang and Huang | MW662661 | ||
| Dichopterinae | Dichoptera sp. | MW662659 | ||
| Pyrops candelaria (Linné) | FJ006724 | |||
| Pyrops clavatus (Westwood) | MW662665 | |||
| Pyrops lathburii (Kirby) | MW662666 | |||
| Pyrops spinolae (Westwood) | MW662667 | |||
| Outgroup | Membracidae | Entylia carinata (Forster) | NC_033539 | |
| Leptobelus gazella Fairmaire | NC_023219 | |||
| Aetalionidae | Darthula hardwickii Gray, | NC_026699 | ||
| Cicadellidae | Drabescoides nuchalis (Jacobi) | NC_028154 | ||
| Nephotettix cincticeps (Uhler) | NC_026977 | |||
| Aphididae | Acyrthosiphon pisum (Harris) | NC_011594 | ||
| Aphalaridae | Pachypsylla venusta (Osten-Sacken) | NC_006157 |
| Species | Regions | Size (bp) | T(U) | C | A | G | AT(%) | GC(%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| D. | PCGs | 10,944 | 43.7 | 12.3 | 33.7 | 10.4 | 77.4 | 22.7 | −0.13 | −0.082 |
| 1st codon position | 3648 | 37.7 | 11.2 | 36.2 | 14.9 | 73.9 | 26.1 | −0.02 | 0.141 | |
| 2nd codon position | 3648 | 49.6 | 18.1 | 19.8 | 12.4 | 69.4 | 30.5 | −0.429 | −0.187 | |
| 3rd codon position | 3648 | 43.8 | 7.4 | 45 | 3.9 | 88.8 | 11.3 | 0.014 | −0.311 | |
| tRNAs | 1422 | 36.3 | 10.1 | 40.2 | 13.4 | 76.5 | 23.5 | 0.051 | 0.14 | |
| rRNAs | 1952 | 49.7 | 8 | 27.6 | 14.7 | 77.3 | 22.7 | −0.287 | 0.296 | |
| A + T-rich region | 1469 | 40.1 | 9 | 43.6 | 7.4 | 83.7 | 16.4 | 0.041 | −0.1 | |
| Full genome | 15,803 | 28.5 | 14.2 | 49.4 | 7.9 | 77.9 | 22.1 | 0.267 | −0.283 | |
| L. | PCGs | 10,956 | 42.5 | 12.5 | 33.9 | 11.2 | 76.4 | 23.7 | −0.112 | −0.056 |
| 1st codon position | 3652 | 36.3 | 11.7 | 36.6 | 15.4 | 72.9 | 27.1 | 0.005 | 0.136 | |
| 2nd codon position | 3652 | 49.4 | 18.5 | 19.6 | 12.6 | 69 | 31.1 | −0.432 | −0.189 | |
| 3rd codon position | 3652 | 41.8 | 7.3 | 45.5 | 5.5 | 87.3 | 12.8 | 0.042 | −0.142 | |
| tRNAs | 1410 | 35.7 | 10.5 | 40.9 | 12.9 | 76.6 | 23.4 | 0.067 | 0.103 | |
| rRNAs | 1945 | 50 | 7.6 | 28.2 | 14.2 | 78.2 | 21.8 | −0.278 | 0.302 | |
| A + T-rich region | 1634 | 34.4 | 9 | 51.2 | 5.4 | 85.6 | 14.4 | 0.196 | −0.246 | |
| Full genome | 15,957 | 27.9 | 14.3 | 49.7 | 8 | 77.6 | 22.3 | 0.281 | −0.282 | |
| N. | PCGs | 10,929 | 42.2 | 12.5 | 33.8 | 11.5 | 76 | 24 | −0.112 | −0.044 |
| 1st codon position | 3643 | 36.1 | 11.3 | 36.6 | 16 | 72.7 | 27.3 | 0.006 | 0.173 | |
| 2nd codon position | 3643 | 47.9 | 18.5 | 20.6 | 13 | 68.5 | 31.5 | −0.398 | −0.176 | |
| 3rd codon position | 3643 | 42.7 | 7.8 | 44.1 | 5.4 | 86.8 | 13.2 | 0.016 | −0.178 | |
| tRNAs | 1410 | 35.7 | 11.2 | 39.9 | 13.3 | 75.6 | 24.5 | 0.055 | 0.084 | |
| rRNAs | 1945 | 48.6 | 8.7 | 28.2 | 14.6 | 76.8 | 23.3 | −0.266 | 0.252 | |
| A + T-rich region | 2082 | 28.7 | 7.9 | 56 | 7.4 | 84.7 | 15.3 | 0.322 | −0.028 | |
| Full genome | 16,510 | 28.6 | 13.4 | 48.7 | 9.3 | 77.3 | 22.7 | 0.261 | −0.18 | |
| Pea. | PCGs | 10,959 | 43.5 | 12.2 | 33.7 | 10.7 | 77.2 | 22.9 | −0.127 | −0.066 |
| 1st codon position | 3653 | 37 | 11 | 36.9 | 15.1 | 73.9 | 26.1 | −0.001 | 0.157 | |
| 2nd codon position | 3653 | 48.8 | 18.4 | 20 | 12.8 | 68.8 | 31.2 | −0.419 | −0.179 | |
| 3rd codon position | 3653 | 44.6 | 7.1 | 44.2 | 4.1 | 88.8 | 11.2 | −0.005 | −0.273 | |
| tRNAs | 1399 | 36.5 | 10.4 | 40.2 | 12.9 | 76.7 | 23.3 | 0.048 | 0.104 | |
| rRNAs | 1945 | 48.8 | 7.9 | 28.3 | 15 | 77.1 | 22.9 | −0.265 | 0.312 | |
| A + T-rich region | 1728 | 34.8 | 10.2 | 48.3 | 6.8 | 83.1 | 17 | 0.162 | −0.201 | |
| Full genome | 16,093 | 29.2 | 13.9 | 48.6 | 8.3 | 77.8 | 22.2 | 0.249 | −0.254 | |
| Pec. | PCGs | 10,953 | 41.6 | 13.7 | 33.4 | 11.4 | 75 | 25.1 | −0.11 | −0.092 |
| 1st codon position | 3651 | 35.3 | 12.4 | 36.3 | 16 | 71.6 | 28.4 | 0.015 | 0.126 | |
| 2nd codon position | 3651 | 48.6 | 18.6 | 19.9 | 13 | 68.5 | 31.6 | −0.42 | −0.177 | |
| 3rd codon position | 3651 | 40.9 | 10.1 | 43.9 | 5.1 | 84.8 | 15.2 | 0.035 | −0.324 | |
| tRNAs | 1428 | 36.1 | 10.9 | 39.5 | 13.5 | 75.6 | 24.4 | 0.045 | 0.106 | |
| rRNAs | 1977 | 48.9 | 7.9 | 28.2 | 15 | 77.1 | 22.9 | −0.269 | 0.307 | |
| A + T-rich region | 1598 | 44.6 | 9.8 | 40.7 | 5 | 85.3 | 14.8 | −0.046 | −0.322 | |
| Full genome | 16,040 | 29.2 | 15.5 | 47.3 | 8.1 | 76.5 | 23.6 | 0.237 | −0.314 | |
| Pev. | PCGs | 10,947 | 43.1 | 12.8 | 33 | 11.1 | 76.1 | 23.9 | −0.132 | −0.071 |
| 1st codon position | 3649 | 36.4 | 11.8 | 35.7 | 16.1 | 72.1 | 27.9 | −0.01 | 0.154 | |
| 2nd codon position | 3649 | 49.7 | 18.3 | 19.3 | 12.7 | 69 | 31 | −0.441 | −0.179 | |
| 3rd codon position | 3649 | 43 | 8.4 | 44 | 4.6 | 87 | 13 | 0.011 | −0.294 | |
| tRNAs | 1422 | 35.9 | 11 | 39.5 | 13.6 | 75.4 | 24.6 | 0.048 | 0.105 | |
| rRNAs | 1960 | 48.4 | 8 | 28.8 | 14.8 | 77.2 | 22.8 | −0.254 | 0.298 | |
| A + T-rich region | 1327 | 36.9 | 12.7 | 43.5 | 6.8 | 80.4 | 19.5 | 0.082 | −0.305 | |
| Full genome | 15,814 | 28.6 | 15 | 48.1 | 8.4 | 76.7 | 23.4 | 0.254 | −0.282 | |
| Pyc. | PCGs | 10,950 | 40.7 | 13.8 | 33.4 | 12.1 | 74.1 | 25.9 | −0.099 | −0.065 |
| 1st codon position | 3650 | 35.1 | 12.1 | 36.3 | 16.5 | 71.4 | 28.6 | 0.017 | 0.156 | |
| 2nd codon position | 3650 | 47.8 | 18.8 | 19.9 | 13.4 | 67.7 | 32.2 | −0.413 | −0.168 | |
| 3rd codon position | 3650 | 39.1 | 10.5 | 43.9 | 6.4 | 83 | 16.9 | 0.057 | −0.242 | |
| tRNAs | 1405 | 35.1 | 11.9 | 38.9 | 14.2 | 74 | 26.1 | 0.051 | 0.087 | |
| rRNAs | 1948 | 48.7 | 8.8 | 27.6 | 14.9 | 76.3 | 23.7 | −0.277 | 0.257 | |
| A + T-rich region | 1611 | 32.3 | 8.5 | 51.9 | 7.3 | 84.2 | 15.8 | 0.232 | −0.079 | |
| Full genome | 16,054 | 26.7 | 15.3 | 48.6 | 9.4 | 75.3 | 24.7 | 0.29 | −0.24 | |
| Pyl. | PCGs | 10,956 | 40.1 | 14.7 | 32.3 | 13 | 72.3 | 27.7 | −0.108 | −0.059 |
| 1st codon position | 3652 | 34.4 | 12.5 | 36.1 | 16.9 | 70.5 | 29.4 | 0.024 | 0.151 | |
| 2nd codon position | 3652 | 47.7 | 19 | 19.9 | 13.4 | 67.6 | 32.4 | −0.412 | −0.171 | |
| 3rd codon position | 3652 | 38 | 12.6 | 40.7 | 8.7 | 78.7 | 21.3 | 0.034 | −0.18 | |
| tRNAs | 1421 | 34.8 | 11.4 | 39.9 | 13.9 | 74.7 | 25.3 | 0.069 | 0.1 | |
| rRNAs | 1943 | 48.5 | 9.6 | 26.5 | 15.4 | 75 | 25 | −0.294 | 0.232 | |
| A + T-rich region | 1672 | 32.2 | 8.7 | 51.3 | 7.8 | 83.5 | 16.5 | 0.228 | −0.051 | |
| Full genome | 16,104 | 26.4 | 15.5 | 47.7 | 10.4 | 74.1 | 25.9 | 0.288 | −0.199 | |
| Pys. | PCGs | 10,959 | 39.6 | 15.3 | 31.8 | 13.2 | 71.4 | 28.5 | −0.109 | −0.073 |
| 1st codon position | 3653 | 34.1 | 12.8 | 36.2 | 17 | 70.3 | 29.8 | 0.03 | 0.141 | |
| 2nd codon position | 3653 | 47.3 | 19.5 | 19.7 | 13.6 | 67 | 33.1 | −0.413 | −0.18 | |
| 3rd codon position | 3653 | 37.4 | 13.7 | 39.7 | 9.2 | 77.1 | 22.9 | 0.03 | −0.197 | |
| tRNAs | 1420 | 34.7 | 11.1 | 39.7 | 14.5 | 74.4 | 25.6 | 0.067 | 0.135 | |
| rRNAs | 1944 | 48.3 | 9.6 | 27 | 15.1 | 75.3 | 24.7 | −0.284 | 0.222 | |
| A + T-rich region | 1595 | 32.8 | 8.8 | 50.8 | 7.6 | 83.6 | 16.4 | 0.215 | −0.069 | |
| Full genome | 16,028 | 26.4 | 15.8 | 47.2 | 10.7 | 73.6 | 26.5 | 0.283 | −0.193 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Zhang, H.; Constant, J.; Bartlett, C.R.; Qin, D. Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha). Genes 2021, 12, 1185. https://doi.org/10.3390/genes12081185
Wang W, Zhang H, Constant J, Bartlett CR, Qin D. Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha). Genes. 2021; 12(8):1185. https://doi.org/10.3390/genes12081185
Chicago/Turabian StyleWang, Wenqian, Huan Zhang, Jérôme Constant, Charles R. Bartlett, and Daozheng Qin. 2021. "Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha)" Genes 12, no. 8: 1185. https://doi.org/10.3390/genes12081185
APA StyleWang, W., Zhang, H., Constant, J., Bartlett, C. R., & Qin, D. (2021). Characterization, Comparative Analysis and Phylogenetic Implications of Mitogenomes of Fulgoridae (Hemiptera: Fulgoromorpha). Genes, 12(8), 1185. https://doi.org/10.3390/genes12081185





