Abstract
The complete mitogenomes of nine fulgorid species were sequenced and annotated to explore their mitogenome diversity and the phylogenetics of Fulgoridae. All species are from China and belong to five genera: Dichoptera Spinola, 1839 (Dichoptera sp.); Neoalcathous Wang and Huang, 1989 (Neoalcathous huangshanana Wang and Huang, 1989); Limois Stål, 1863 (Limois sp.); Penthicodes Blanchard, 1840 (Penthicodes atomaria (Weber, 1801), Penthicodes caja (Walker, 1851), Penthicodes variegata (Guérin-Méneville, 1829)); Pyrops Spinola, 1839 (Pyrops clavatus (Westwood, 1839), Pyrops lathburii (Kirby, 1818), Pyrops spinolae (Westwood, 1842)). The nine mitogenomes were 15,803 to 16,510 bp in length with 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs) and a control region (A + T-rich region). Combined with previously reported fulgorid mitogenomes, all PCGs initiate with either the standard start codon of ATN or the nonstandard GTG. The TAA codon was used for termination more often than the TAG codon and the incomplete T codon. The nad1 and nad4 genes varied in length within the same genus. A high percentage of F residues were found in the nad4 and nad5 genes of all fulgorid mitogenomes. The DHU stem of trnV was absent in the mitogenomes of all fulgorids sequenced except Dichoptera sp. Moreover, in most fulgorid mitogenomes, the trnL2, trnR, and trnT genes had an unpaired base in the aminoacyl stem and trnS1 had an unpaired base in the anticodon stem. The similar tandem repeat regions of the control region were found in the same genus. Phylogenetic analyses were conducted based on 13 PCGs and two rRNA genes from 53 species of Fulgoroidea and seven outgroups. The Bayesian inference and maximum likelihood trees had a similar topological structure. The major results show that Fulgoroidea was divided into two groups: Delphacidae and ((Achilidae + (Lophopidae + (Issidae + (Flatidae + Ricaniidae)))) + Fulgoridae). Furthermore, the monophyly of Fulgoridae was robustly supported, and Aphaeninae was divided into Aphaenini and Pyropsini, which includes Neoalcathous, Pyrops, Datua Schmidt, 1911, and Saiva Distant, 1906. The genus Limois is recovered in the Aphaeninae, and the Limoisini needs further confirmation; Dichoptera sp. was the earliest branch in the Fulgoridae.
1. Introduction
The family Fulgoridae is one of the large groups in the superfamily Fulgoroidea (Hemiptera: Auchenorrhyncha), consisting of 142 genera and 774 species [1], distributed worldwide but mainly in pan-tropical regions [1,2]. Many fulgorid species are brilliantly colored with an elongate and often strangely shaped head process. Some produce cuticular waxes, comprised mostly of keto esters in a variety of forms [3], including plumes that can extend well beyond the length of the abdomen (e.g., in Cerogenes auricoma (Burmeister, 1835)) [4]. Some fulgorids are agricultural pests, such as Lycorma delicatula (White, 1845); both nymphs and adults may cause direct feeding damage and indirect damage to plants through sooty mold growth on excreted honeydew [5,6].
The family Fulgoridae requires further assessment to obtain a robust phylogenetic assessment and resultant classificatory system [7]. The monophyly of the Aphaeninae, the second-largest subfamily of Fulgoridae comprising 35 genera and 207 species distributed mostly in the Old World [1] remains doubtful [7]. Moreover, the genus Pyrops Spinola, a large and showy genus of 69 species of tropical Asia and the Indomalayan region, remains of incertae sedis at the subfamily level [8]. Furthermore, the placement and status of Dichopterinae remain controversial [9,10,11,12,13,14]. More evidence, including mitogenomes, are needed to address these problems in Fulgoridae.
Here, we presented the complete sequenced and annotated mitogenomes of nine fulgorid species. We compared these mitogenomes with those previously published among Fulgoroidea and combined available mitogenomes to explore the diversity of mitogenomes and phylogenetics of Fulgoroidea, with particular reference to Fulgoridae.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
Specimens of nine fulgorid species were collected from China. All specimens were preserved in 100% ethanol and stored at −20 °C in the Entomological Museum of the Northwest A&F University. After morphological identification, the thoracic muscle tissue was used to extract total genomic DNA using the DNeasy DNA Extraction kit (Qiagen). Species identifications were based on Melichar (1912) [15] for the genus Dichoptera, Wang et al. (2020) [16] for the genus Limois, Constant and Pham (2018) [17] for the genus Neoalcathous, Constant (2010) [18] for the genus Penthicodes and Nagai and Porion (1996) [19] for the genus Pyrops.
The collection data of the extracted specimens are as follows:
Specimens of Dichoptera sp. were collected by Lijia Wang from Hainan Province, Mt. Jianfengling in August 2020 (Figure 1A); Limois sp. were collected by Daozheng Qin from Shaanxi Province, Huoditang in August 2019 (Figure 1B); Neoalcathous huangshanana Wang and Huang, 1989, was collected by Deliang Xu from Guangdong Province, Mt. Nanling in August 2020 (Figure 1C); Penthicodes atomaria (Weber, 1801) were collected by Wenqian Wang from Hainan Province, Mt. Diaoluo in April 2018 (Figure 1D); Penthicodes caja (Walker, 1851) were collected by Wenqian Wang from Yunnan Province, Guanping in April 2019 (Figure 1E); Penthicodes variegata (Guérin-Méneville, 1829) were collected by Wenqian Wang from Yunnan Province, Mengla in April 2019 (Figure 1F); Pyrops clavatus (Westwood, 1839) were collected by Wenqian Wang from Guangxi Province, Debao, Hongfeng forest park in June 2018 (Figure 1G); Pyrops lathburii (Kirby, 1818) were collected by Na Zhang from Guangxi Province in July 2017 (Figure 1H); Pyrops spinolae (Westwood, 1842) were collected by Wenqian Wang from Guangxi Province, Pingxiang in June 2018 (Figure 1I).
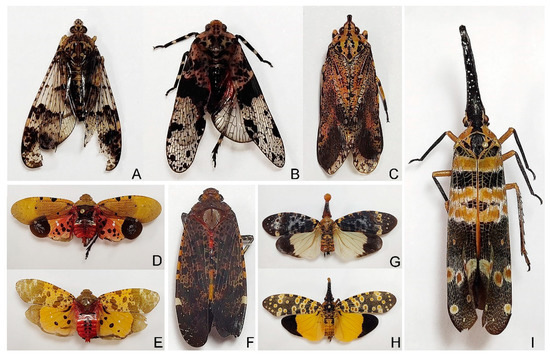
Figure 1.
Photo plate of nine Fulgoridae specimens.
2.2. Mitogenomes Sequence Analysis
The whole mitogenomes of the nine species were sequenced using next-generation sequencing (NGS) (Illumina NovaSeq platform with paired-ends of 2 × 150 bp). Quality triming and assembly of raw paired reads were performed by Geneious 11.0.2 (Biomatters, Auckland, New Zealand) with default parameters [20]. The mitochondrial genome of L. delicatula (EU909203), Pyrops candelaria (Linné, 1758) (FJ006724), Aphaena (Callidepsa) amabilis (Hope, 1843) (MN025522) and Aphaena (Aphaena) discolor nigrotibiata Schmidt, 1906 (MN025523) (Hemiptera: Fulgoridae) [21,22] were chosen as bait sequences. Geneious 11.0.2 was used for mitogenomes annotation with L. delicatula, Py. candelaria, A. (C.) amabilis and A. (A.) discolor nigrotibiata (Hemiptera: Fulgoridae) used as references. All 13 PCGs were identified by open reading frames and translated into amino acids under the invertebrate mitochondrial genetic code. MITOS WebServer was run to identify and predict secondary structures of 22 tRNAs [23]. The secondary structures of tRNAs were mapped by Adobe Illustrator CS5 manually. After aligning with other mitogenomes in Fulgoridae, the rRNA genes and control region were annotated by the boundary of the adjacent tRNA genes. The tandem repeats of the control region were identified by the tandem repeats finder online server [24]. CGView was used to produce the whole mitogenome map [25]. PhyloSuite was performed to analyze the base composition and relative synonymous codon usage (RSCU) [26]. The sliding window analysis of 200 bp in a step size of 20 bp was performed with DnaSP v 5.0 to estimate nucleotide diversity (Pi) base on 13 PCGs and 2 rRNA genes from fourteen fulgorid mitogenomes (nine in this paper and five previously reported). Then, the DnaSP v5 was used to calculate the nucleotide diversity (Pi) of each PCG and rRNA [27].
2.3. Phylogenetic Analysis
Phylogenetic analyses were performed based on 13 PCGs and two rRNA genes among 53 species in Fulgoroidea, including 27 species in Delphacidae, 3 species in Ricaniidae, 2 species in Issidae, 1 species in Flatidae, 1 species in Lophopidae, 5 species in Achilidae and 14 species in Fulgoridae. The outgroups included five species from Membracoidea and two species from Sternorrhyncha. All the mitogenomes used in this study can be searched from GenBank (Table 1). Each PCG gene was aligned individually in codon-alignment mode under MAFFT and the results were concatenated by PhyloSuite [26]. Codon alignment modes were aligned using the G-INS-i (accurate) strategy, and RNA sequences were aligned using the Q-INS-i strategy normal alignment mode. Ambiguous sites and gaps in the alignments were removed using Gblocks v0.91b [28]. The optimal nucleotide substitution models and partition strategies were selected by PartitionFinder2. The best-fitting model was used for each partition with the greedy search algorithm and Bayesian information criterion (BIC) [26]. Four different datasets were concatenated for analyses: PCG123R matrix (all three codon positions of PCGs and two rRNA genes); PCG123 matrix (all three codon positions of PCGs); PCG12R matrix (the first and second codon positions of PCGs and two rRNA genes); PCG12 matrix (the first and second codon positions of PCGs). The maximum likelihood (ML) and Bayesian inference (BI) were employed based on four datasets for phylogenetic analyses. The maximum likelihood analyses were conducted by IQ-TREE [29], under an ML + rapid bootstrap (BS) algorithm with 1000 replicates. The Bayesian analyses were performed by MrBayes 3.2.6 [30] with two simultaneous runs of four chains each. Markov Chain Monte Carlo (MCMC) sampling estimated the posterior distributions using the settings for 5 × 106 MCMC generations, with a relative burn-in of 25%, and MCMC termination when the average standard deviation of split frequencies fell below 0.01. PhyloBayes MPI v1.5a was used for phylogenetic reconstruction of the four datasets based on the site-heterogeneous model CAT + GTR on CIPRES [31,32]. Two independent trees were searched and the analysis was terminated when the two runs reached convergence (maxdiff below 0.3 and minimum effective size above 50). The initial 25% of each MCMC chain run was discarded as burn-in and a consensus tree was generated from the remaining trees combined from two runs.

Table 1.
The mitogenomic sequences used in this study.
3. Results
3.1. Mitogenome Organization and Gene Content
The complete mitogenome sequence of Dichoptera sp. was 15,803 bp, Limois sp. was 15,957 bp, N. huangshanana was 16,510 bp, Pe. atomaria was 16,093 bp, Pe. caja was 16,040 bp, Pe. variegata was 15,814 bp, Py. clavatus was 16,054 bp, Py. lathburii was 16,104 bp and Py. spinolae was 16,028 bp in length, respectively (Figure 2). Variations in the length of mitogenomes may be owing to a variable number of repeats in the control regions. The mitogenomes of nine fulgorid species encode 37 genes (13 PCGs, two rRNAs, and 22 tRNAs) and a control region (A + T-rich region) (Figure 2; Supplementary Tables S1–S3). All nine sequences exhibited the uniform gene arrangement as other planthopper mitogenomes. The nine mitogenomes displayed a heavy AT nucleotide bias, with an A + T% range from 73.6 to 77.9%: 77.9% in Dichoptera sp., 77.6% in Limois sp., 77.3% in N. huangshanana, 77.8% in Pe. atomaria, 76.5% in Pe. caja, 76.7% in Pe. variegata, 75.3% in Py. clavatus, 74.1% in Py. lathburii and 73.6% in Py. spinolae. This is moderate compared to levels found in other planthopper species. After comparing these nine mitogenomes, the composition skew analysis showed that all nine fulgorid species present a positive AT skew and a negative GC skew in the whole mitogenome and had a lower A + T content in tRNAs than in rRNAs. The control region (Table 2) has a positive AT skew and a negative GC skew in all nine mitogenome sequences except Pe. caja, which shows a negative AT skew and GC skew. A conserved overlap of 7 bp between atp8 and atp6 was found in nine sequences (Figure 3).
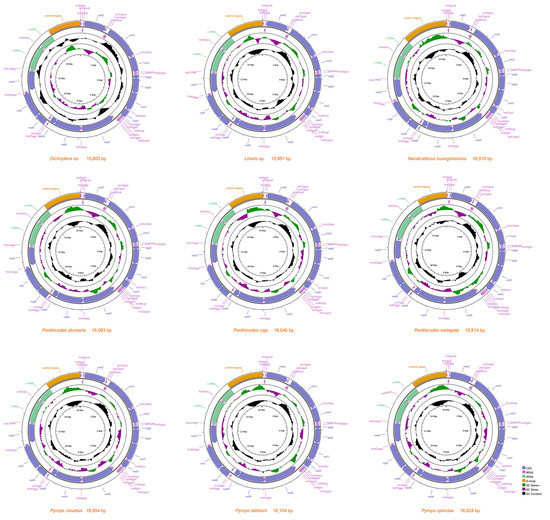
Figure 2.
Organization of nine Fulgoridae complete mitogenomes.

Table 2.
Nucleotide composition of the mitogenomes of nine Fulgoridae.
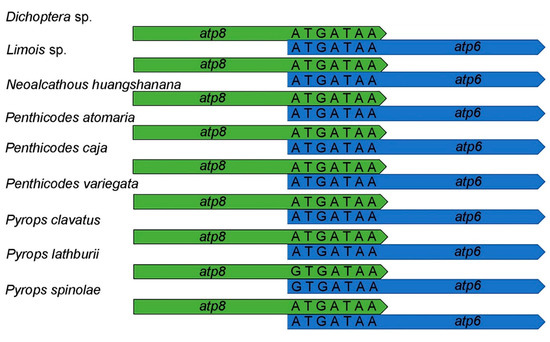
Figure 3.
Sequence alignments of atp8-atp6.
3.2. Protein-Coding Genes and Relative Synonymous Codon Usage
The total size of the 13 PCGs of nine mitogenomes ranged between 10,929 and 10,959 bp. In all nine mitogenomes, PCGs showed negative AT skew and GC skew. The A + T content of the third codon position was much higher than of the first and the second positions. The third codon position had a positive AT skew and negative GC skew in all nine species except Pe. atomaria, which had a negative AT skew and GC skew (Table 2). Most PCGs started with the codon ATN, except for atp6 and nad1 in Py. lathburii, which initiated with the codon GTG (Supplementary Tables S1–S4). This had also been found in other Fulgoroidea mitogenomes [21,33]. Comparing the nine sequenced mitogenomes, the result showed that atp6 terminated with an incomplete T codon; nad2, nad6, cox1, cox2 and atp8 ended with a complete TAA codon; nad5 ended with a TAN codon except for Pe. caja and Pe. variegata using the incomplete stop codon T. Transcribed truncated stop codons might be converted to TAA by polyadenylation [34]. In all nine sequenced mitogenomes, the ATG codon was used more often for starting than other codons and the GTG codon was the least used (Supplementary Tables S1–S4). The TAA codon appeared more often than the TAG codon, and the incomplete T codon was the least used in Fulgoridae. After comparing those sequences, we found nad1 and nad4 varied in length among species in the same genus (Supplementary Tables S1–S3). A high percentage of F residues appeared in nad4 and nad5 in all nine mitogenome sequences near the start position in nad4, and both the start and end position in nad5.
The result of relative synonymous codon usage (RSCU) (Figure 4) of the nine fulgorids shows that the codon usage of all genes had a strong bias toward the nucleotides A and T, particularly the third codon positions. Leucine (Leu), Phenylalanine (Phe), Isoleucine (Ile), Methionine (Met) and Serine (Ser) were used most frequently in the nine fulgorid mitogenomes sequenced. In addition, the codons Pro (CCG) and Thr (ACG) were absent in Dichoptera sp. The codons Arg (CGC) and Ser1 (AGC) were absent in Limois sp. The codons Pro (CCG) and Arg (CGC) deficiency was found in Pe. atomaria. The codon Arg (CGC) was not observed in Py. clavatus.
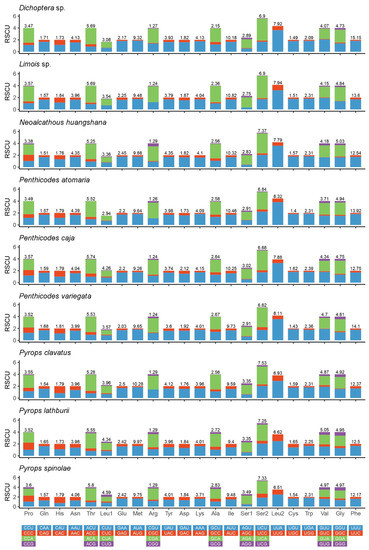
Figure 4.
Relative synonymous codon usage (RSCU) in the mitogenomes of nine fulgorid species.
3.3. Transfer and Ribosomal RNA Genes
A total of 22 tRNA genes and two rRNA genes were identified in the same relative genomic positions as those observed for the previously sequenced fulgorid genomes [21,22]. The tRNAs had a positive AT skew and GC skew in all nine fulgorid species (Table 2). The total lengths of 22 tRNAs were 1399 to 1428 bp and 1943 to 1977 bp in rRNA (Table 2). The trnV was with a reduced DHU arm in all nine species except Dichoptera sp. (Supplementary Figures S1–S9). All nine fulgorid species were missing the DHU stem of this trnS1. The TΨC of trnE arms was missing in Dichoptera sp. All tRNA genes of the nine fulgorid species were the highly conserved, structures of 7 bp in the aminoacyl stem, 7 bp in the anticodon loop, and 5 bp in the anticodon stem, while the length of the DHU and TΨC arms was variable. In addition, trnT, trnR, and trnL2 had an unpaired base in the aminoacyl stem in all nine fulgorid mitogenomes except Dichoptera sp., whose trnT were without an unpaired base in the aminoacyl stem. The anticodon stem of trnS1 had an unpaired base in N. huangshanana, Pe. atomaria and Py. clavatus. Six types of unmatched base pairs (G-U, U-U, A-A, G-A, A-C and U-C) were found in the nine mitogenomes. The rRNAs had a negative AT skew and positive GC skew in nine sequenced mitogenomes (Table 2).
The large RNA genes were located between trnL1 and trnV, with the sizes ranging from 1210 to 1224 bp, and the small ribosomal RNA genes were located between trnV and the control region, with the sizes ranging from 730 to 756 bp. The rRNA genes showed a heavy AT nucleotide bias, with A + T content 77.3% in Dichoptera sp., 78.2% in Limois sp., 76.8% in N. huangshanana, 77.1% in Pe. atomaria, 77.1% in Pe. caja, 77.2% in Pe. variegata, 76.3% in Py. clavatus, 75% in Py. lathburii and 75.3% in Py. spinolae, which were similar to those found in other sequenced Fulgoridae (Figure 2; Supplementary Tables S1–S3, Table 2).
3.4. Control Region
The putative control region (A + T-rich region) was annotated at the conserved position between rrnS and trnI (Table 2). Except for Pe. caja with negative AT skew and GC skew, all nine fulgorid species showed positive AT skew and negative GC skew in the control region. In the nine sequenced mitogenomes, the size of this region was 1327 bp to 2082 bp, and the A + T content of this region (more than 80%) was one of the highest within the whole mitogenome sequence (Table 2). One tandem repeat region was found in the control region of Py. lathburii, Py. clavatus, Py. spinolae, Pe. caja, Pe. variegata and Dichoptera sp.; Pe. atomaria, N. huangshanana and Limois sp. had two tandem repeat regions. The repeat unit of “TGCAAAAAAA(A)” was found in Py. lathburii, Py. clavatus, Py. spinolae and N. huangshanana. The repeat unit of “TTGCAAAAAA(A)” was found in Pe. caja and Pe. variegata, while a similar repeat unit of “TTGCAAAAA(A/T)(A)” was found in Pe. atomaria; TT/GCAAAAAAA(A) as the second repeat unit was found in N. huangshanana. Limois sp. has the repeat unit “TCATAAAAAA(A)”. The same repeat unit “TTGCAAAAAA(A)” was also found in two species, A. (C.) amabilis and A. (A.) discolor nigrotibiata in the genus Aphaena Guérin-Méneville, 1834 [22]. Moreover, 1–3 Poly(A) or Poly(T) could be found in those fulgorid species (Figure 5). Interestingly, a Poly(A) was consistently located near the 3′-end of the control regions in all the sequenced Fulgoridae species except Pe. caja, Pe. variegata and N. huangshanana (Figure 5).
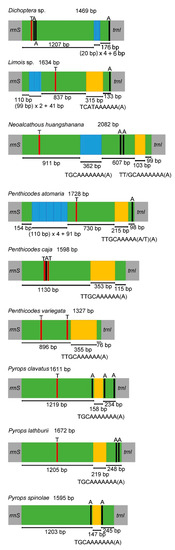
Figure 5.
Structures of the control regions in the nine Fulgoridae mitogenomes. The blue and yellow boxes show the tandem repeats (the uppercase below represents the repeat units of sequences). The green boxes are non-repeat regions. The black and red blocks were the structures of poly (A) and poly (T), respectively.
3.5. Nucleotide Diversity
The results of nucleotide diversity based on 13 PCGs and two rRNA genes from fourteen sequenced Fulgoridae species (Figure 6) show that nucleotide diversity values range from 0.136 (rrnL and rrnS) to 0.264 (atp8). Comparing each gene, atp8 (Pi = 0.264) presents the highest variability, followed by nad2 (Pi = 0.240) and nad6 (Pi = 0.238). However, cox1 (Pi = 0.155) and nad1 (Pi = 0.155) were the comparatively conserved genes in the 13 PCGs. Two rRNA genes were also highly conserved, with lower values of 0.136 in rrnL and rrnS, respectively.
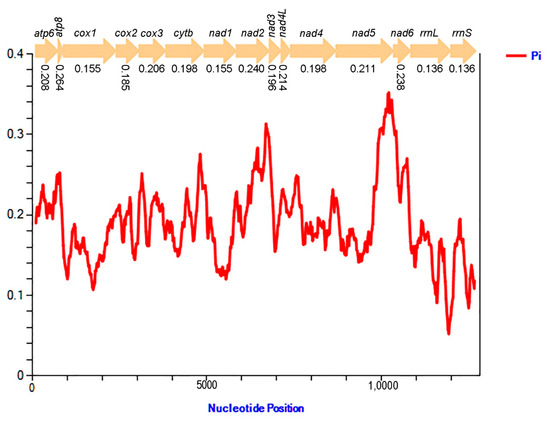
Figure 6.
Sliding window analyses based on 13 PCGs and two rRNA genes of fourteen Fulgoridae mitogenomes. The red line indicates the value of nucleotide diversity (Pi) (window of 200 bp with the step size of 20 bp). The Pi value of each gene is below the gene name.
3.6. Phylogenetic Analyses
Under the best models (Supplementary Table S5) selected by PartitionFinder and the heterogeneous model CAT + GTR, the ML and BI analyses based on PCG123R, PCG123, PCG12R and PCG12 datasets yielded similar tree topologies (Figure 7 and Supplementary Figures S10–S19), Bayesian posterior probabilities (PP) and bootstrap values (BS), all of which are shown on individual nodes. Overall, most received moderate to high support (Bayesian PP = 0.9–1/BS = 70–100) [35,36]. BI analysis and ML analysis provided better resolution with stronger support values. The results supported the monophyly of the lineages of the superfamily Fulgoroidea. The families Delphacidae, Achilidae, Ricaniidae, and Fulgoridae were also monophyletic in our analyses. Meanwhile, the results show that Fulgoroidea was divided into two groups: Delphacidae and ((Achilidae + (Lophopidae + (Issidae + (Flatidae + Ricaniidae)))) + Fulgoridae) with the PP = 1 and BS = 100, except the tree produced based on the PCG12R dataset under the CAT + GTR model, which was ((Lophopidae + (Issidae + (Flatidae + Ricaniidae))) + (Achilidae+ Fulgoridae)). The phylogenetic analysis also supported the early branching of the family Delphacidae with high support values (PP/BS = 1/100), supporting the previous studies of Urban and Cryan (2007) and Song and Liang (2013) [37,38]. Furthermore, the hypothetical relationships of (Lophopidae + (Issidae + (Flatidae + Ricaniidae))) in the superfamily Fulgoroidea were robustly supported. Within Fulgoridae, the phylogenetic relationships indicated that Aphaeninae was divided into two clades that we consider as representing the tribe Aphaenini Blanchard, 1847 (genera Aphaena Guérin-Méneville, 1834; Limois Stål, 1863; Lycorma Stål, 1863; Penthicodes Blanchard, 1845) and Pyropsini Urban and Cryan, 2009 (genera Neoalcathous Wang and Huang, 1989; Pyrops Spinola, 1839); Dichoptera sp. was branching off earlier than other species in Fulgoridae; the genus Lycorma was the sister to Aphaena (Figure 7); the tribe Limoisini is not recovered.
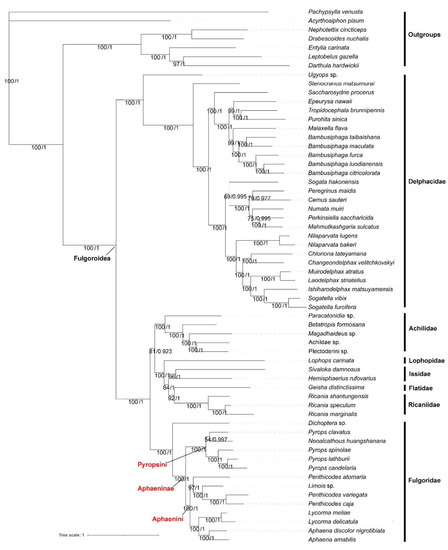
Figure 7.
Phylogenetic tree inferred from ML and BI (MrBayes) analysis based on PCG123R matrix. Bootstrap values (BS) and Bayesian posterior probabilities (PP) are indicated on branches.
4. Discussion
4.1. Comparative Analysis of Fulgorid Mitogenomes
The nine sequenced fulgorid mitogenomes were conservative and similar to other species of Fulgoroidea from previous studies [33,39,40,41,42,43,44,45,46]. Their length ranged from 15,803 to 16,510 bp, comparable to the previously reported fulgorid mitogenomes that ranged from 15,946 bp (in L. delicatula) to 16,237 bp (in A. (C.) amabilis) [21,22,46]. These different lengths were mainly attributed to the varied size of intergenic spacer regions and the length of AT-rich regions.
The nine fulgorid mitogenomes exhibited an extremely high A + T content, ranging from 77.9% in Dichoptera sp. to 73.6% in Py. spinolae. These values are comparable to the previously reported fulgorid mitogenomes which ranged from 74.3% (in Py. candelaria) to 77.9% (in A. (C.) amabilis) [21,22,46]. The region at nad3-nad4l was extremely high in A + T content, serial tRNAs and stable stem-loop structures which may disrupt PCR and sequencing reactions; meanwhile, this study also found the poly (A) in nad4 and nad5, presenting a high percentage of F residues. All these may cause the mitogenome sequences of most fulgorid species to become rather difficult. This phenomenon was also found in some other Hemiptera species [21,22,46,47,48,49,50].
Wang et al. (2019) suggested that a missing DHU stem in the trnV of Aphaena was unusual in Fulgoridae. Nevertheless, current evidence suggests that the mitogenomes of fulgorid species all lack stable DHU stems in trnV except Dichoptera sp. (Supplementary Figures S1–S9) [21,22,46], which made the missing DHU stem in trnV seem a common feature in Fulgoridae.
The size of the A + T-rich region in the nine sequenced species was similar to other Fulgoroidea, except Sivaloka damnosus Chow and Lu (in Issidae) [41]. Here, we found diversity in the repeat unit in Fulgoridae. The genus Penthicodes had a similar repeat unit as that found in Aphaena and Lycorma, and the same repeat unit was also found in Pyrops and N. huangshanana, but the repeat unit of Dichoptera sp. is remarkably different from the other sequenced species. Tandem repeats were also identified in the control region of the mitogenomes in several families in Fulgoroidea, including Ricaniidae, Flatidae, Delphacidae, Issidae, Fulgoridae and Achilidae [21,22,33,39,41,42,44,45]. The control region of Fulgoroidea presented a dramatic divergence in the base composition, fragment length and the repeat units.
4.2. Nucleotide Diversity of Fulgorid Mitogenomtes
Mitochondrial genes have been widely used as genetic markers, especially the cox1 gene, including widespread use as DNA barcodes for identifying species and testing phylogenetic relationships among related taxa [51]. However, cox1 remains inefficient for species delimitation in some groups owing to intra- and interspecific variations [52,53,54]. This analysis indicates that the cox1 gene is relatively conserved and exhibits a slow evolution rate. The nad1 gene is also highly conserved, while atp8, nad2 and nad6 genes have relatively faster evolution rates. Although atp8 with 165 bp may be too short to be phylogenetically informative, nad2 and nad6 could be selected as potential DNA markers to clarify the phylogenetic relationships of closely related species of Fulgoridae.
4.3. Phylogeny and Species Delimitation
The genus Pyrops is endemic to the Indomalayan Region, which indubitably belongs to the Old World lineages, as indicated in the two alternative biogeographic hypotheses of Urban and Cryan (2009) [7]. Wang and Huang (1989) [55] erected the genus Neoalcathous and placed it in the subfamily Amyclinae based on its similarities to Alcathous Stål.
However, this placement of Oriental genera in a subfamily based on Neotropical taxa was shown to be erroneous by the subsequent molecular study of Urban and Cryan (2009) [7], and Neoalcathous was hence transferred to the subfamily Aphaeninae by Constant and Pham (2018) [17]. In the current study, N. huangshanana was placed in a clade with the genus Pyrops. The morphological characters also show similarities between N. huangshanana and the genus Pyrops in being larger; cephalic process curved dorsally; pronotum with median carina (sometimes obsolete) and a small but strongly impressed point on each side; mesonotum with median carina; tegmina colored, at most 3 times as long as broad, with the apical margin more or less rounded [17,55,56]. Here, we hypothesize that N. huangshanana is closely related to the genus Pyrops and we place them in a tribe Pyropsini Urban and Cryan, 2009, together with the closely related Saiva Distant, 1906, and Datua Schmidt, 1911 [7,57], in the subfamily Aphaeninae. The genus Limois apparently belongs to the tribe Aphaenini, and the tribe Limoisini might need to be considered a synonym of the latter in the future. However, further study including additional genera placed in Limoisini (Bloeteanella Lallemand, 1959, Erilla Distant, 1906, Neolieftinckana Lallemand, 1963, Nisax Fennah, 1977, Ombro Fennah, 1977, Saramel Fennah, 1977) is necessary to refine and confirm or refute the relevance of the tribe Limoisini.
Dichoptera sp. belongs to the tribe Dichopterini in the Dichopterinae. However, the assignment of the subfamily Dichopterinae was once controversial [13,14]. This study found trnT with a paired base in the aminoacyl stem and a lack of stable DHU stem in trnV in Dichoptera sp., which is obviously different from other fulgorid species. However, more evidence is still needed before ascertaining the status and placement of Dichopterinae, including the addition of representatives of Dictyopharidae into phylogenetic studies.
5. Conclusions
The nine fulgorid mitogenomes which belong to five genera (Dichoptera Spinola, 1839; Neoalcathous Wang and Huang, 1989; Limois Stål, 1863; Penthicodes Blanchard, 1840 and Pyrops Spinola, 1839) were sequenced. The arrangement of nine sequenced fulgorid mitogenomes are conservative and similar to other species of Fulgoridae. The comprehensive analysis can provide a more profound understanding of the mitochondrial characteristics among Fulgoridae.
The phylogenetic analyses showed that Delphacidae was the sister to ((Achilidae + (Lophopidae + (Issidae + (Flatidae + Ricaniidae)))) + Fulgoridae). The subfamily Aphaeni-nae was divided into Aphaenini and Pyropsini. The genus Limois is recovered in the Aphaeninae, apparently belongs to the tribe Aphaenini, and the Limoisini might need to be considered a synonym which needs further confirmation. Dichoptera sp. was the earliest branch in the Fulgoridae. Compared with other fulgorid mitogenomes, Dichoptera sp. has obvious difference in trnT and trnV. Ascertaining the status and placement of Dichopterinae still needed more study to do. Increasing the sample and mitochondrial data to solve the problems existing in the phylogeny of Fulgoridae is our future work.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12081185/s1, Figure S1. Secondary structure for the tRNAs of Dichoptera sp.; Figure S2. Secondary structure for the tRNAs of Limois sp.; Figure S3. Secondary structure for the tRNAs of N. huangshanana; Figure S4. Secondary structure for the tRNAs of Pe. atomaria; Figure S5. Secondary structure for the tRNAs of Pe. caja; Figure S6. Secondary structure for the tRNAs of Pe. variegata; Figure S7. Secondary structure for the tRNAs of Py. clavatus; Figure S8. Secondary structure for the tRNAs of Py. lathburii; Figure S9. Secondary structure for the tRNAs of Py. spinolae; Figure S10. ML tree based on PCG123. BS are indicated on branches; Figure S11. BI (MrBayes) tree based on PCG123. PP are indicated on branches; Figure S12. ML tree based on PCG12R. BS are indicated on branches; Figure S13. BI (MrBayes) tree based on PCG12R. PP are indicated on branches; Figure S14. ML tree based on PCG12. BS are indicated on branches; Figure S15. BI (MrBayes) tree based on PCG12. PP are indicated on branches; Figure S16. BI (PhyloBayes) tree based on PCG123R. PP are indicated on branches; Figure S17. BI (PhyloBayes) tree based on PCG123. PP are indicated on branches; Figure S18. BI (PhyloBayes) tree based on PCG12R. PP are indicated on branches; Figure S19. BI (PhyloBayes) tree based on PCG12. PP are indicated on branches; Table S1. Mitogenomic organization of nine Fulgoridae mitochondrial genomes. Dichoptera sp. (D.); Limois sp. (L.); Neoalcathous huangshanana (N.); Table S2. Mitogenomic organization of nine Fulgoridae mitochondrial genomes. Penthicodes atomaria (Pea.); Penthicodes caja (Pec.); Penthicodes variegata (Pev.); Table S3. Mitogenomic organization of nine Fulgoridae mitochondrial genomes. Pyrops clavatus (Pyc.); Pyrops lathburii (Pyl.) and Pyrops spinolae (Pys.); Table S4. Start and stop codons usage of nine Fulgoridae mitogenomes; Table S5. Best partitioning scheme and models used in this study.
Author Contributions
Investigation, formal analysis, and writing—original draft, W.W.; resources, H.Z.; writing—review and editing, W.W., J.C.; C.R.B. and D.Q.; and validation, project administration, and funding acquisition: D.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (grant numbers 31672340, 31750002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers 31672340, 31750002).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bourgoin, T. FLOW (Fulgoromorpha Lists on the Web), Version 8. Available online: http://hemipteradatabases.org/flow/ (accessed on 6 June 2021).
- Metcalf, Z.P. General Catalogue of the Hemiptera; Fasc. IV: Fulgoroidea. Part. 9: Fulgoridae; North Carolina State College: Raleigh, NC, USA, 1947. [Google Scholar]
- Mason, R.T.; Fales, H.M.; Jones, T.H.; O’Brien, L.B.; Taylor, T.W.; Hogue, C.L.; Blum, M.S. Characterization of fulgorid waxes (Homoptera: Fulgoroidea: Insecta). Insect Biochem. 1989, 19, 737–740. [Google Scholar] [CrossRef]
- Goemans, G. The Fulgoridae (Hemiptera, Fulgoromorpha) of Guatemala. In Biodiversidad de Guatemala; Cano, E.B., Ed.; The Universidad del Valle de Guatemala: Guatemala City, Guatemala, 2006; Volume 1, pp. 337–344. [Google Scholar]
- Lee, D.H.; Park, Y.L.; Leskey, T.C. A review of biology and management of Lycorma delicatula (Hemiptera: Fulgoridae), an emerging global invasive species. J. Asia Pac. Entomol. 2019, 22, 589–596. [Google Scholar] [CrossRef]
- Urban, J.M. Perspective: Shedding light on spotted lanternfly impacts in the USA. Pest Manag. Sci. 2019, 76, 10–17. [Google Scholar] [CrossRef]
- Urban, J.M.; Cryan, J.R. Entomologically famous, evolutionarily unexplored: The first phylogeny of the lanternfly family Fulgoridae (Insecta: Hemiptera: Fulgoroidea). Mol. Phylogenetics Evol. 2009, 50, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Constant, J.; Pham, H.T. Review of the clavatus group of the lanternfly genus Pyrops (Hemiptera: Fulgoromorpha: Fulgoridae). Eur. J. Taxon. 2017, 305, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Kirkaldy, G.W. Bibliographical and nomenclatorial notes on the Hemiptera No. 3. Entomologist 1904, 37, 279–283. [Google Scholar] [CrossRef]
- Muir, F.A.G. On the classification of the Fulgoroidea (Homoptera). In Proceedings of the Hawaiian Entomological Society, Honolulu, HI, USA, 6 April 1922; Volume 5, pp. 205–247. [Google Scholar]
- Metcalf, Z.P. The Fulgorina of Barro Colorado and other parts of Panama. In Bulletin of the Museum of Comparative Zoology at Harvard College; The Museum: Cambridge, MA, USA, 1938; Volume 82, pp. 277–423. [Google Scholar] [CrossRef]
- Emeljanov, A.F. The problem of differentiation of the families Fulgoridae and Dictyopharidae. Tr. Zool. Inst. Akad. Nauk SSSR 1979, 82, 3–22. [Google Scholar]
- Emeljanov, A.F. Improved Tribal Delimitation of the Subfamily Dictyopharinae and Description of New Genera and New Species (Homoptera, Fulgoroidea, Dictyopharidae). Entomol. Rev. 2011, 90, 299–328. [Google Scholar] [CrossRef]
- Emeljanov, A.F. A new species of the genus Pibrocha Kirkaldy (Homoptera, Fulgoridae) with notes on the systematics of the subfamily Dichopterinae and with description of a new tribe. Entomol. Rev. 2013, 92, 123–129. [Google Scholar] [CrossRef]
- Melichar, L. Monographie der Dictyophorinen (Homoptera). Abh. K. K. Zool. Bot. Ges. Wien Wien 1912, 7, 1–221. [Google Scholar]
- Wang, W.Q.; Xu, S.L.; Constant, J.; Qin, D.Z. Revision of the lanternfly genus Limois Stål, 1863 (Hemiptera: Fulgoromorpha: Fulgoridae) with description of a new species from China. Eur. J. Taxon. 2021, 720, 35–61. [Google Scholar] [CrossRef]
- Constant, J.; Pham, H.T. Extension of the lanternfly genus Neoalcathous Wang and Huang, 1989 to Vietnam with a new species and new subfamily placement (Hemiptera: Fulgoromorpha: Fulgoridae). Belg. J. Entomol. 2018, 59, 1–11. [Google Scholar]
- Constant, J. The lanternfly genus Penthicodes: Key to the species and review of the “Ereosoma group” with two new species and one new subspecies (Hemiptera: Fulgoromorpha: Fulgoridae). Zootaxa 2010, 2523, 1–26. [Google Scholar] [CrossRef]
- Nagai, S.; Porion, T. Fulgoridae 2: Catalogue Illustré des Faunes Asiatique et Australienne; Sciences Nat: Compiègne, France, 1996; p. 80. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Havas, S.S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liang, A.P.; Bu, C.P. A Molecular Phylogeny of Hemiptera Inferred from Mitochondrial Genome Sequences. PLoS ONE 2012, 7, e48778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.Q.; Huang, Y.X.; Bartlett, C.R.; Zhou, F.M.; Meng, R.; Qin, D.Z. Characterization of the complete mitochondrial genomes of two species of the genus Aphaena Guérin-Méneville (Hemiptera: Fulgoridae) and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 141, 29–40. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.V.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Mark, P.V.D.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.; Pfeiffer, W.T.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; Institute of Electrical and Electronics Engineers (IEEE): New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Huang, Y.X.; Qin, D.Z. Sequencing and analysis of the complete mitochondrial genome of Changeondelphax velitchkovskyi (Hemiptera: Fulgoroidea). Mitochondrial DNA Part B 2018, 3, 90–91. [Google Scholar] [CrossRef] [Green Version]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondrial. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Bui, M. IQ-TREE Manual: Frequently Asked Questions (WWW Document). Available online: http://www.iqtree.org/doc/Frequently-Asked-Questions (accessed on 23 March 2021).
- Urban, J.M.; Cryan, J.R. Evolution of the planthoppers (Insecta: Hemiptera: Fulgoroidea). Mol. Phylogenetics Evol. 2007, 42, 556–572. [Google Scholar] [CrossRef]
- Song, N.; Liang, A.P. A preliminary molecular phylogeny of planthoppers (Hemiptera: Fulgoroidea) based on nuclear and mitochondrial DNA sequences. PLoS ONE 2013, 8, e58400. [Google Scholar] [CrossRef] [Green Version]
- Song, N.; Liang, A.P. The complete mitochondrial genome sequence of Geisha distinctissima (Hemiptera: Flatidae) and comparison with other hemipteran insects. Acta Biochim. Biophys. Sin. 2009, 41, 206–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Liang, A.P. Complete mitochondrial genome of the small brown planthopper, Laodelphax striatellus (Delphacidae: Hemiptera), with a novel gene order. Zool. Sci. 2009, 26, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liang, A.P.; Ma, C. The complete mitochondrial genome sequence of the planthopper, Sivaloka damnosus. J. Insect Sci. 2010, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.J.; Zhu, W.C.; Rong, X.; Ding, X.L.; Zhang, Y.K.; Liu, J.; Chen, D.S.; Du, Y.; Hong, X.Y. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: Conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genom. 2013, 14, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.; Peng, X.X.; Jing, S.L.; Liu, B.F.; Zhu, L.L.; He, G.C. Intraspecific and interspecific variations in the mitochondrial genomes of Nilaparvata (Hemiptera: Delphacidae). J. Econ. Entomol. 2015, 108, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.X.; Guan, D.L.; Niu, Y.; Sang, L.Q.; Zhang, X.X.; Xu, S.Q. Characterization of the complete mitochondrial genome of the Asian planthopper Ricania speculum (Hemiptera: Fulgoroidea: Ricannidae). Conserv. Genet. Resour. 2016, 8, 463–466. [Google Scholar] [CrossRef]
- Xu, S.Y.; Long, J.K.; Chen, X.S. Comparative analysis of the complete mitochondrial genomes of five Achilidae species (Hemiptera: Fulgoroidea) and other Fulgoroidea reveals conserved mitochondrial genome organization. PeerJ 2019, 7, e6659. [Google Scholar] [CrossRef]
- Du, Z.Y.; Wu, Y.F.; Chen, Z.; Cao, L.M.; Ishikawa, T.; Kamitani, S.; Sota, T.; Song, F.; Tian, L.; Cai, W.Z.; et al. Global Phylogeography and Invasion History of the Spotted Lanternfly Revealed by Mitochondrial Phylogenomics. Evol. Appl. 2020, 14, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, A.; Nobandegani, F.F. Detection and evaluation of hydrogen bond strength in nucleic acid base pairs. J. Phys. Chem. A 2008, 112, 281–295. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, X.L.; Qiao, G.X. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE 2013, 8, e77511. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, W.L.; Claessens, A.; Otto, T.D.; Kekre, M.; Fairhurst, R.M.; Rayner, J.C.; Kwiatkowski, D. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017, 45, 1889–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Huang, X.; Deng, J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020, 158, 854–864. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Lunt, D.H.; Zhang, D.X.; Szymura, J.M.; Hewltt, O.M. The insect cytochrome oxidase I gene: Evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol. Biol. 1996, 5, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Shiyang, K.; Vaidya, G.; Ng, P.K.L. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Whitworth, T.L.; Dawson, R.D.; Magalon, H.; Baudry, E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc. R. Soc. B Biol. Sci. 2007, 274, 1731–1739. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Huang, J. Notes of new genus and species of Fulgoroidea from China (Homoptera: Fulgoroidea) (2). Acta Agric. Boreali Sin. 1989, S1, 127–132. [Google Scholar]
- Constant, J. Review of the effusus group of the lanternfly genus Pyrops Spinola, 1839, with one new species and notes on trophobiosis (Hemiptera: Fulgoromorpha: Fulgoridae). Eur. J. Taxon. 2015, 128, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Lallemand, V. Revision des Fulgoridae (Homoptera). Deuxième partie. Faunes Asiatique et Australienne. In Mémoires de l’Institut Royal des Sciences Naturelles de Belgique; Deuxième Série; Koninklijk Belgisch instituut voor Natuur Wetenschappen: Brussel, Belgisch, 1963; Volume 75, pp. 1–99. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).