Host Genetics and Gut Microbiome: Perspectives for Multiple Sclerosis
Abstract
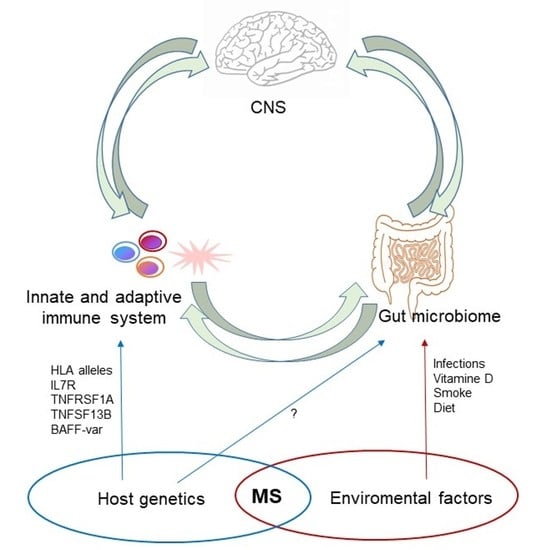
1. Introduction
2. Genetic Factors in MS Patients
3. The Gut Microbiome and MS
3.1. The Human Gut Microbiota
3.2. Factors Influencing the Composition of Gut Microbiota
3.3. Gut Microbiota-Immune-Brain Interactions
3.4. Evidences from Experimental Autoimmune Encephalomyelitis (EAE)
3.5. Evidences from Clinical Studies in MS Patients
4. Interactions between Gut Microbiota and Host Genes
4.1. Heritability of Gut Microbiome
4.2. MGWAS Studies
4.3. Evidence from Autoimmune Diseases and MS
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Dutta, R.; Trapp, B.D. Mechanisms of neuronal dysfunction and degeneration in multiple sclerosis. Prog. Neurobiol. 2011, 93, 1–12. [Google Scholar] [CrossRef]
- Wallin, M.T.; Culpepper, W.J.; Nichols, E.; Bhutta, Z.A.; Gebrehiwot, T.T.; Hay, S.I.; Khalil, I.A.; Krohn, K.J.; Liang, X.; Naghavi, M.; et al. Global, regional, and national burden of multiple sclerosis 1990–2016: A systematic analysis for the Global Burden of Disease Study. Lancet Neurol. 2019, 18, 269–285. [Google Scholar] [CrossRef]
- Voskuhl, R.R. The effect of sex on multiple sclerosis risk and disease progression. Mult. Scler. J. 2020, 26, 554–560. [Google Scholar] [CrossRef]
- Fish, E.N. The X-files in immunity: Sex-based differences predispose immune responses. Nat. Rev. Immunol. 2008, 8, 737–744. [Google Scholar] [CrossRef]
- Voci, C. Testicular hypofunction and multiple sclerosis: Cause or consequence? Ann. Neurol. 2014, 76, 765. [Google Scholar] [CrossRef]
- Durelli, L.; Conti, L.; Clerico, M.; Boselli, D.; Contessa, G.; Ripellino, P.; Ferrero, B.; Eid, P.; Novelli, F. T-helper 17 cells expand in multiple sclerosis and are inhibited by interferon-β. Ann. Neurol. 2009, 65, 499–509. [Google Scholar] [CrossRef]
- Rolla, S.; Bardina, V.; De Mercanti, S.; Quaglino, P.; De Palma, R.; Gned, D.; Brusa, D.; Durelli, L.; Novelli, F.; Clerico, M. Th22 cells are expanded in multiple sclerosis and are resistant to IFN-β. J. Leukoc. Biol. 2014, 96, 1155–1164. [Google Scholar] [CrossRef]
- Costantino, C.M.; Baecher-Allan, C.; Hafler, D.A. Multiple Sclerosis and Regulatory T Cells. J. Clin. Immunol. 2008, 28, 697–706. [Google Scholar] [CrossRef]
- Langrish, C.L.; Chen, Y.; Blumenschein, W.M.; Mattson, J.; Basham, B.; Sedgwick, J.D.; McClanahan, T.; Kastelein, R.A.; Cua, D.J. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005, 201, 233–240. [Google Scholar] [CrossRef]
- Kebir, H.; Kreymborg, K.; Ifergan, I.; Dodelet-Devillers, A.; Cayrol, R.; Bernard, M.; Giuliani, F.; Arbour, N.; Becher, B.; Prat, A. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 2007, 13, 1173–1175. [Google Scholar] [CrossRef]
- Tzartos, J.S.; Friese, M.A.; Craner, M.J.; Palace, J.; Newcombe, J.; Esiri, M.M.; Fugger, L. Interleukin-17 Production in Central Nervous System-Infiltrating T Cells and Glial Cells Is Associated with Active Disease in Multiple Sclerosis. Am. J. Pathol. 2008, 172, 146–155. [Google Scholar] [CrossRef]
- Voet, S.; Prinz, M.; van Loo, G. Microglia in Central Nervous System Inflammation and Multiple Sclerosis Pathology. Trends Mol. Med. 2019, 25, 112–123. [Google Scholar] [CrossRef]
- Schirmer, L.; Schafer, D.P.; Bartels, T.; Rowitch, D.H.; Calabresi, P.A. Diversity and Function of Glial Cell Types in Multiple Sclerosis. Trends Immunol. 2021, 42, 228–247. [Google Scholar] [CrossRef]
- Westerlind, H.; Ramanujam, R.; Uvehag, D.; Kuja-Halkola, R.; Boman, M.; Bottai, M.; Lichtenstein, P.; Hillert, J. Modest familial risks for multiple sclerosis: A registry-based study of the population of Sweden. Brain 2014, 137, 770–778. [Google Scholar] [CrossRef]
- Sadovnick, A.D.; Armstrong, H.; Rice, G.P.A.; Bulman, D.; Hashimoto, L.; Party, D.W.; Hashimoto, S.A.; Warren, S.; Hader, W.; Murrary, T.J.; et al. A population-based study of multiple sclerosis in twins: Update. Ann. Neurol. 2004, 33, 281–285. [Google Scholar] [CrossRef]
- Hollenbach, J.A.; Oksenberg, J.R. The immunogenetics of multiple sclerosis: A comprehensive review. J. Autoimmun. 2015, 64, 13–25. [Google Scholar] [CrossRef]
- Sawcer, S.; Hellenthal, G.; Pirinen, M.; Spencer, C.C.A.; Patsopoulos, N.A.; Moutsianas, L.; Dilthey, A.; Su, Z.; Freeman, C.; Hunt, S.E.; et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476, 214–219. [Google Scholar] [CrossRef]
- Belbasis, L.; Bellou, V.; Evangelou, E.; Ioannidis, J.P.A.; Tzoulaki, I. Environmental risk factors and multiple sclerosis: An umbrella review of systematic reviews and meta-analyses. Lancet Neurol. 2015, 14, 263–273. [Google Scholar] [CrossRef]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Handunnetthi, L.; Giovannoni, G.; Ramagopalan, S.V. An Updated Meta-Analysis of Risk of Multiple Sclerosis following Infectious Mononucleosis. PLoS ONE 2010, 5, e12496. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, R.; Li, Z.; Wang, Y.; Gao, C.; Lv, X.; Song, Y.; Li, B. The risk of smoking on multiple sclerosis: A meta-analysis based on 20,626 cases from case-control and cohort studies. PeerJ 2016, 4, e1797. [Google Scholar] [CrossRef]
- McDowell, T.-Y.; Amr, S.; Culpepper, W.J.; Langenberg, P.; Royal, W.; Bever, C.; Bradham, U.D. Sun Exposure, Vitamin D Intake and Progression to Disability among Veterans with Progressive Multiple Sclerosis. Neuroepidemiology 2011, 37, 52–57. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Budhram, A.; Parvathy, S.; Kremenchutzky, M.; Silverman, M. Breaking down the gut microbiome composition in multiple sclerosis. Mult. Scler. J. 2016, 23, 628–636. [Google Scholar] [CrossRef]
- Amini, M.E.; Shomali, N.; Bakhshi, A.; Rezaei, S.; Hemmatzadeh, M.; Hosseinzadeh, R.; Eslami, S.; Babaie, F.; Aslani, S.; Torkamandi, S.; et al. Gut microbiome and multiple sclerosis: New insights and perspective. Int. Immunopharmacol. 2020, 88, 107024. [Google Scholar] [CrossRef]
- Mumford, C.J.; Wood, N.W.; Kellar-Wood, H.; Thorpe, J.W.; Miller, D.H.; Compston, D.A. The British Isles Survey of Multiple Sclerosis in Twins. Neurology 1994, 44, 11–15. [Google Scholar] [CrossRef]
- Hansen, T.; Skytthe, A.; Stenager, E.; Petersen, H.C.; Kyvik, K.O.; Brønnum-Hansen, H. Risk for Multiple Sclerosis in Dizygotic and Monozygotic Twins. Mult. Scler. 2005, 11, 500–503. [Google Scholar] [CrossRef]
- Lincoln, M.R.; Montpetit, A.; Cader, M.Z.; Saarela, J.; Dyment, D.A.; Tiislar, M.; Ferretti, V.; Tienari, P.J.; Sadovnick, A.D.; Peltonen, L.; et al. A Predominant Role for the HLA Class II Region in the Association of the MHC Region with Multiple Sclerosis. Nat. Genet. 2005, 37, 1108–1112. [Google Scholar] [CrossRef]
- Goris, A.; Pauwels, I.; Dubois, B. Progress in Multiple Sclerosis Genetics. Curr. Genom. 2012, 13, 646–663. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium (IMSGC); Beecham, A.H.; Patsopoulos, N.A.; Xifara, D.K.; Davis, M.F.; Kemppinen, A.; Cotsapas, C.; Shah, T.S.; Spencer, C.; Booth, D.; et al. Analysis of Immune-Related Loci Identifies 48 New Susceptibility Variants for Multiple Sclerosis. Nat. Genet. 2013, 45, 1353–1360. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium Multiple Sclerosis Genomic Map Implicates Peripheral Immune Cells and Microglia in Susceptibility. Science 2019, 365, 7188. [CrossRef] [PubMed]
- Gregory, S.G.; Schmidt, S.; Seth, P.; Oksenberg, J.R.; Hart, J.; Prokop, A.; Caillier, S.J.; Ban, M.; Goris, A.; Barcellos, L.F.; et al. Interleukin 7 Receptor Alpha Chain (IL7R) Shows Allelic and Functional Association with Multiple Sclerosis. Nat. Genet. 2007, 39, 1083–1091. [Google Scholar] [CrossRef]
- Galarza-Muñoz, G.; Briggs, F.B.S.; Evsyukova, I.; Schott-Lerner, G.; Kennedy, E.M.; Nyanhete, T.; Wang, L.; Bergamaschi, L.; Widen, S.G.; Tomaras, G.D.; et al. Human Epistatic Interaction Controls IL7R Splicing and Increases Multiple Sclerosis Risk. Cell 2017, 169, 72–84.e13. [Google Scholar] [CrossRef]
- Gregory, A.P.; Dendrou, C.A.; Attfield, K.E.; Haghikia, A.; Xifara, D.K.; Butter, F.; Poschmann, G.; Kaur, G.; Lambert, L.; Leach, O.A.; et al. TNF Receptor 1 Genetic Risk Mirrors Outcome of Anti-TNF Therapy in Multiple Sclerosis. Nature 2012, 488, 508–511. [Google Scholar] [CrossRef]
- Steri, M.; Orrù, V.; Idda, M.L.; Pitzalis, M.; Pala, M.; Zara, I.; Sidore, C.; Faà, V.; Floris, M.; Deiana, M.; et al. Overexpression of the Cytokine BAFF and Autoimmunity Risk. N. Engl. J. Med. 2017, 376, 1615–1626. [Google Scholar] [CrossRef]
- Mitrovič, M.; Patsopoulos, N.A.; Beecham, A.H.; Dankowski, T.; Goris, A.; Dubois, B.; D’Hooghe, M.B.; Lemmens, R.; Van Damme, P.; Søndergaard, H.B.; et al. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell 2018, 175, 1679–1687.e7. [Google Scholar] [CrossRef]
- Zuk, O.; Hechter, E.; Sunyaev, S.R.; Lander, E.S. The Mystery of Missing Heritability: Genetic Interactions Create Phantom Heritability. Proc. Natl. Acad. Sci. USA 2012, 109, 1193–1198. [Google Scholar] [CrossRef]
- Wu, G.D.; Lewis, J.D. Analysis of the Human Gut Microbiome and Association with Disease. Clin. Gastroenterol. Hepatol. 2013, 11, 774–777. [Google Scholar] [CrossRef]
- Mielcarz, D.W.; Kasper, L.H. The Gut Microbiome in Multiple Sclerosis. Curr. Treat. Options Neurol. 2015, 17, 344. [Google Scholar] [CrossRef]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human Microbiome: An Academic Update on Human Body Site Specific Surveillance and Its Possible Role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef] [PubMed]
- Fricker, A.M.; Podlesny, D.; Fricke, W.F. What Is New and Relevant for Sequencing-Based Microbiome Research? A Mini-Review. J. Adv. Res. 2019, 19, 105–112. [Google Scholar] [CrossRef]
- Hugon, P.; Dufour, J.-C.; Colson, P.; Fournier, P.-E.; Sallah, K.; Raoult, D. A Comprehensive Repertoire of Prokaryotic Species Identified in Human Beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef]
- Shvets, Y.V.; Lukianova, N.Y.; Chekhun, V.F. Human Microbiota and Effectiveness of Cancer Chemotherapy. Exp. Oncol. 2020, 42, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of Short-Chain Fatty Acid Production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the Normal Gut Microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Gur, T.L. Intrauterine Microbiota: Missing, or the Missing Link? Trends Neurosci. 2019, 42, 402–413. [Google Scholar] [CrossRef]
- Moore, R.E.; Townsend, S.D. Temporal Development of the Infant Gut Microbiome. Open Biol. 2019, 9, 190128. [Google Scholar] [CrossRef]
- Dobbler, P.; Mai, V.; Procianoy, R.S.; Silveira, R.C.; Corso, A.L.; Roesch, L.F.W. The Vaginal Microbial Communities of Healthy Expectant Brazilian Mothers and Its Correlation with the Newborn’s Gut Colonization. World J. Microbiol. Biotechnol. 2019, 35, 159. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- McLoughlin, K.; Schluter, J.; Rakoff-Nahoum, S.; Smith, A.L.; Foster, K.R. Host Selection of Microbiota via Differential Adhesion. Cell Host Microbe 2016, 19, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.-D.; Wang, Y.-D. The Roles of the Gut Microbiota–MiRNA Interaction in the Host Pathophysiology. Mol. Med. 2020, 26, 101. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Luckey, D.; Taneja, V. The Gut Microbiome in Autoimmunity: Sex Matters. Clin. Immunol. 2015, 159, 154–162. [Google Scholar] [CrossRef]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef]
- Davis, E.C.; Dinsmoor, A.M.; Wang, M.; Donovan, S.M. Microbiome Composition in Pediatric Populations from Birth to Adolescence: Impact of Diet and Prebiotic and Probiotic Interventions. Dig. Dis. Sci. 2020, 65, 706–722. [Google Scholar] [CrossRef]
- Trasande, L.; Blustein, J.; Liu, M.; Corwin, E.; Cox, L.M.; Blaser, M.J. Infant Antibiotic Exposures and Early-Life Body Mass. Int. J. Obes. 2013, 37, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the Microbiota, Immune and Nervous Systems in Health and Disease. Nat. Neurosci. 2017, 20, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Mowat, A.M. Anatomical Basis of Tolerance and Immunity to Intestinal Antigens. Nat. Rev. Immunol. 2003, 3, 331–341. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Kadowaki, A.; Quintana, F.J. The Gut-CNS Axis in Multiple Sclerosis. Trends Neurosci. 2020, 43, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.O.; Ochoa-Repáraz, J. The Gut Microbiome in Multiple Sclerosis: A Potential Therapeutic Avenue. Med. Sci. 2018, 6, 69. [Google Scholar] [CrossRef]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagu. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The Microbiome and Innate Immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Huus, K.E.; Petersen, C.; Finlay, B.B. Diversity and Dynamism of IgA−microbiota Interactions. Nat. Rev. Immunol. 2021, 21, 514–525. [Google Scholar] [CrossRef]
- Liang, S.C.; Tan, X.-Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 Are Coexpressed by Th17 Cells and Cooperatively Enhance Expression of Antimicrobial Peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef]
- Cao, A.T.; Yao, S.; Gong, B.; Elson, C.O.; Cong, Y. Th17 Cells Upregulate Polymeric Ig Receptor and Intestinal IgA and Contribute to Intestinal Homeostasis. J. Immunol. 2012, 189, 4666–4673. [Google Scholar] [CrossRef]
- Flannigan, K.L.; Ngo, V.L.; Geem, D.; Harusato, A.; Hirota, S.A.; Parkos, C.A.; Lukacs, N.W.; Nusrat, A.; Gaboriau-Routhiau, V.; Cerf-Bensussan, N.; et al. IL-17A-Mediated Neutrophil Recruitment Limits Expansion of Segmented Filamentous Bacteria. Mucosal Immunol. 2017, 10, 673–684. [Google Scholar] [CrossRef]
- Barnes, M.J.; Powrie, F. Regulatory T Cells Reinforce Intestinal Homeostasis. Immunity 2009, 31, 401–411. [Google Scholar] [CrossRef]
- Neumann, C.; Blume, J.; Roy, U.; Teh, P.P.; Vasanthakumar, A.; Beller, A.; Liao, Y.; Heinrich, F.; Arenzana, T.L.; Hackney, J.A.; et al. C-Maf-Dependent Treg Cell Control of Intestinal TH17 Cells and IgA Establishes Host-Microbiota Homeostasis. Nat. Immunol. 2019, 20, 471–481. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The Microbiota in Adaptive Immune Homeostasis and Disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Berer, K.; Mues, M.; Koutrolos, M.; Rasbi, Z.A.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal Microbiota and Myelin Autoantigen Cooperate to Trigger Autoimmune Demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-Cell Responses to Gut Microbiota Promote Experimental Autoimmune Encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4615–4622. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Guan, N.L.; et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed]
- Shih, V.F.-S.; Cox, J.; Kljavin, N.M.; Dengler, H.S.; Reichelt, M.; Kumar, P.; Rangell, L.; Kolls, J.K.; Diehl, L.; Ouyang, W.; et al. Homeostatic IL-23 Receptor Signaling Limits Th17 Response through IL-22-Mediated Containment of Commensal Microbiota. Proc. Natl. Acad. Sci. USA 2014, 111, 13942–13947. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. Central Nervous System Demyelinating Disease Protection by the Human Commensal Bacteroides Fragilis Depends on Polysaccharide A Expression. J. Immunol. 2010, 185, 4101–4108. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Wang, Y.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. A Polysaccharide from the Human Commensal Bacteroides Fragilis Protects against CNS Demyelinating Disease. Mucosal. Immunol. 2010, 3, 487–495. [Google Scholar] [CrossRef]
- Haghikia, A.; Jörg, S.; Duscha, A.; Berg, J.; Manzel, A.; Waschbisch, A.; Hammer, A.; Lee, D.-H.; May, C.; Wilck, N.; et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 2016, 44, 951–953. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut Bacteria from Multiple Sclerosis Patients Modulate Human T Cells and Exacerbate Symptoms in Mouse Models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple Sclerosis Patients Have a Distinct Gut Microbiota Compared to Healthy Controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the Human Gut Microbiome in Multiple Sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut Microbiota in Multiple Sclerosis: Possible Influence of Immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut Microbiota from Multiple Sclerosis Patients Enables Spontaneous Autoimmune Encephalomyelitis in Mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef]
- Takewaki, D.; Suda, W.; Sato, W.; Takayasu, L.; Kumar, N.; Kimura, K.; Kaga, N.; Mizuno, T.; Miyake, S.; Hattori, M.; et al. Alterations of the Gut Ecological and Functional Microenvironment in Different Stages of Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 22402–22412. [Google Scholar] [CrossRef]
- Rumah, K.R.; Linden, J.; Fischetti, V.A.; Vartanian, T. Isolation of Clostridium Perfringens Type B in an Individual at First Clinical Presentation of Multiple Sclerosis Provides Clues for Environmental Triggers of the Disease. PLoS ONE 2013, 8, e76359. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Rezende, R.M.; Moreira, T.G.; Tankou, S.K.; Cox, L.M.; Wu, M.; Song, A.; Dhang, F.H.; Wei, Z.; Costamagna, G.; et al. Oral Administration of MiR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia Muciniphila. Cell Host Microbe 2019, 26, 779–794.e8. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-Scale Association Analyses Identify Host Factors Influencing Human Gut Microbiome Composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Awany, D.; Allali, I.; Dalvie, S.; Hemmings, S.; Mwaikono, K.S.; Thomford, N.E.; Gomez, A.; Mulder, N.; Chimusa, E.R. Host and Microbiome Genome-Wide Association Studies: Current State and Challenges. Front. Genet. 2018, 9, 637. [Google Scholar] [CrossRef]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human Genetic Variation and the Gut Microbiome in Disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Kevans, D.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Panaccione, R.; Otley, A.; et al. Association of Host Genome with Intestinal Microbial Composition in a Large Healthy Cohort. Nat. Genet. 2016, 48, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru Mdcm, K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host Genetic Variation Impacts Microbiome Composition across Human Body Sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef]
- Davenport, E.R.; Cusanovich, D.A.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Genome-Wide Association Studies of the Human Gut Microbiota. PLoS ONE 2015, 10, e0140301. [Google Scholar] [CrossRef]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-Wide Association Analysis Identifies Variation in Vitamin D Receptor and Other Host Factors Influencing the Gut Microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The Effect of Host Genetics on the Gut Microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R.; Franzosa, E.A.; Rahnavard, G.; Hall, A.B.; Vlamakis, H.; Stevens, C.; Daly, M.J.; Xavier, R.J.; Huttenhower, C. Host Genetic Variation and Its Microbiome Interactions within the Human Microbiome Project. Genome Med. 2018, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Rühlemann, M.C.; Hermes, B.M.; Bang, C.; Doms, S.; Moitinho-Silva, L.; Thingholm, L.B.; Frost, F.; Degenhardt, F.; Wittig, M.; Kässens, J.; et al. Genome-Wide Association Study in 8956 German Individuals Identifies Influence of ABO Histo-Blood Groups on Gut Microbiome. Nat. Genet. 2021, 53, 147–155. [Google Scholar] [CrossRef]
- Goris, A.; Liston, A. The Immunogenetic Architecture of Autoimmune Disease. Cold Spring Harb. Perspect. Biol. 2012, 4, a007260. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.S.; Shedlock, A.M.; Langefeld, C.D. Genetics of Autoimmune Diseases: Insights from Population Genetics. J. Hum. Genet. 2015, 60, 657–664. [Google Scholar] [CrossRef]
- Larosa, M.; Zen, M.; Gatto, M.; Jesus, D.; Zanatta, E.; Iaccarino, L.; Inês, L.; Doria, A. IL-12 and IL-23/Th17 Axis in Systemic Lupus Erythematosus. Exp. Biol. Med. 2019, 244, 42–51. [Google Scholar] [CrossRef]
- Yang, P.; Qian, F.-Y.; Zhang, M.-F.; Xu, A.-L.; Wang, X.; Jiang, B.-P.; Zhou, L.-L. Th17 Cell Pathogenicity and Plasticity in Rheumatoid Arthritis. J. Leukoc. Biol. 2019, 106, 1233–1240. [Google Scholar] [CrossRef]
- Ueno, A.; Jeffery, L.; Kobayashi, T.; Hibi, T.; Ghosh, S.; Jijon, H. Th17 Plasticity and Its Relevance to Inflammatory Bowel Disease. J. Autoimmun. 2018, 87, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Piper, C.J.M.; Matei, D.E.; Blair, P.A.; Rendeiro, A.F.; Orford, M.; Alber, D.G.; Krausgruber, T.; Catalan, D.; Klein, N.; et al. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab. 2020, 31, 837–851.e10. [Google Scholar] [CrossRef] [PubMed]
- Dupraz, L.; Magniez, A.; Rolhion, N.; Richard, M.L.; Da Costa, G.; Touch, S.; Mayeur, C.; Planchais, J.; Agus, A.; Danne, C.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Regulate IL-17 Production by Mouse and Human Intestinal Γδ T Cells. Cell Rep. 2021, 36, 109332. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carrio, J.; López, P.; Sánchez, B.; González, S.; Gueimonde, M.; Margolles, A.; de Los Reyes-Gavilán, C.G.; Suárez, A. Intestinal Dysbiosis Is Associated with Altered Short-Chain Fatty Acids and Serum-Free Fatty Acids in Systemic Lupus Erythematosus. Front. Immunol. 2017, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The Microbiome and Inflammatory Bowel Disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef]
- Jianzhong, H. The Genetic Predisposition and the Interplay of Host Genetics and Gut Microbiome in Crohn Disease. Clin. Lab. Med. 2014, 34, 763–770. [Google Scholar] [CrossRef][Green Version]
- Simmons, A. Crohn’s Disease: Genes, Viruses and Microbes. Nature 2010, 466, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Hamm, C.M.; Gulati, A.S.; Sartor, R.B.; Chen, H.; Wu, X.; Zhang, T.; Rohlf, F.J.; Zhu, W.; Gu, C.; et al. Inflammatory Bowel Diseases Phenotype, C. difficile and NOD2 Genotype Are Associated with Shifts in Human Ileum Associated Microbial Composition. PLoS ONE 2012, 7, e26284. [Google Scholar] [CrossRef]
- Zhang, T.; DeSimone, R.A.; Jiao, X.; Rohlf, F.J.; Zhu, W.; Gong, Q.Q.; Hunt, S.R.; Dassopoulos, T.; Newberry, R.D.; Sodergren, E.; et al. Host Genes Related to Paneth Cells and Xenobiotic Metabolism Are Associated with Shifts in Human Ileum-Associated Microbial Composition. PLoS ONE 2012, 7, e30044. [Google Scholar] [CrossRef]
- Gampa, A.; Engen, P.A.; Shobar, R.; Mutlu, E.A. Relationships between Gastrointestinal Microbiota and Blood Group Antigens. Physiol. Genom. 2017, 49, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Ley, R.E.; Clark, A.G. ABO Antigen and Secretor Statuses Are Not Associated with Gut Microbiota Composition in 1500 Twins. BMC Genom. 2016, 17, 941. [Google Scholar] [CrossRef]
- Wells, P.M.; Williams, F.M.K.; Matey-Hernandez, M.L.; Menni, C.; Steves, C.J. ‘RA and the Microbiome: Do Host Genetic Factors Provide the Link? J. Autoimmun. 2019, 99, 104–115. [Google Scholar] [CrossRef]
- Yamamoto, E.A.; Jørgensen, T.N. Relationships Between Vitamin D, Gut Microbiome, and Systemic Autoimmunity. Front. Immunol. 2019, 10, 3141. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zeng, B.; Zhang, J.; Li, W.; Mou, F.; Wang, H.; Zou, Q.; Zhong, B.; Wu, L.; Wei, H.; et al. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci. Rep. 2016, 6, 30594. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-C.; Chun, S.; Kim, K.; Mak, A. Update on the Genetics of Systemic Lupus Erythematosus: Genome-Wide Association Studies and Beyond. Cells 2019, 8, 1180. [Google Scholar] [CrossRef]
- He, Z.; Shao, T.; Li, H.; Xie, Z.; Wen, C. Alterations of the Gut Microbiome in Chinese Patients with Systemic Lupus Erythematosus. Gut Pathog. 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- van der Meulen, T.A.; Harmsen, H.J.M.; Vila, A.V.; Kurilshikov, A.; Liefers, S.C.; Zhernakova, A.; Fu, J.; Wijmenga, C.; Weersma, R.K.; de Leeuw, K.; et al. Shared Gut, but Distinct Oral Microbiota Composition in Primary Sjögren’s Syndrome and Systemic Lupus Erythematosus. J. Autoimmun. 2019, 97, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Hevia, A.; Milani, C.; López, P.; Cuervo, A.; Arboleya, S.; Duranti, S.; Turroni, F.; González, S.; Suárez, A.; Gueimonde, M.; et al. Intestinal Dysbiosis Associated with Systemic Lupus Erythematosus. mBio 2014, 5, e01548-14. [Google Scholar] [CrossRef] [PubMed]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a Gut Pathobiont Drives Autoimmunity in Mice and Humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- Korach-Rechtman, H.; Freilich, S.; Gerassy-Vainberg, S.; Buhnik-Rosenblau, K.; Danin-Poleg, Y.; Bar, H.; Kashi, Y. Murine Genetic Background Has a Stronger Impact on the Composition of the Gut Microbiota than Maternal Inoculation or Exposure to Unlike Exogenous Microbiota. Appl. Environ. Microbiol. 2019, 85, 18. [Google Scholar] [CrossRef]
- Buhnik-Rosenblau, K.; Danin-Poleg, Y.; Kashi, Y. Predominant Effect of Host Genetics on Levels of Lactobacillus Johnsonii Bacteria in the Mouse Gut. Appl. Environ. Microbiol. 2011, 77, 6531–6538. [Google Scholar] [CrossRef]
- Montgomery, T.L.; Künstner, A.; Kennedy, J.J.; Fang, Q.; Asarian, L.; Culp-Hill, R.; D’Alessandro, A.; Teuscher, C.; Busch, H.; Krementsov, D.N. Interactions between Host Genetics and Gut Microbiota Determine Susceptibility to CNS Autoimmunity. Proc. Natl. Acad. Sci. USA 2020, 117, 27516–27527. [Google Scholar] [CrossRef]
- Cahana, I.; Iraqi, F.A. Impact of Host Genetics on Gut Microbiome: Take-Home Lessons from Human and Mouse Studies. Anim. Models Exp. Med. 2020, 3, 229–236. [Google Scholar] [CrossRef]
| Observation/Microorganism | Host Organisms | Possible MS-Related Immunopathogenetic Mechanism | Reference |
|---|---|---|---|
| Germ-free gut | EAE mice | Milder disease course; lower levels of IFN-γ and IL-17A in the gut and CNS; Treg expanded in the CNS; disrupted BBB tight junctions. | [72] [73] [74] |
| Segmented Filamentous bacteria (SFB) | EAE mice | Higher levels of IL-17 in the gut; Th17 expanded in the CNS | [73] |
| Bacteroides fragilis | EAE mice | Polysaccharide A-mediated Tregs induction via TLR2 and suppression of the Th17 response | [76,77] |
| Decreased abundance of Clostridia [79], Bacteroides and Parabacteroides [79,80,81], Butyricimonas [82], Faecalibacterium [83], Prevotella [79,80,81], Lactobacillus [81], Adlercreutzia and Collinsella [81] with respect to controls | MS patients | SCFA-producer bacteria induce IL-10 dependent enhanced conversion of Treg cells and may be related to MS pathogenesis and progression; Clostridia can induce colon regulatory T cells (Tregs) and prevent autoimmunity; Epsilon toxin (ETX) produced by Clostridium perfringens has tropism for the blood-brain barrier (BBB) and oligodendrocytes/myelin. | [24,25] [85] [79] [86] |
| Increased abundance of Metanobrevibacter [82], Acinetobacter calcoaceticus [80,84], Akkermansia muciniphila [80,82,84], Pseudomonas, Mycoplana, Blautia and Dorea [81] with respect to controls | MS patients | Methanogenic archaea can induce DCs activation; Akkermansia and Acinetobacter have been associated with lower Treg induction and increased Th1 polarization. MS fecal transplant induced higher incidence of spontaneous EAE. | [24] [80,82,84] |
| Study | Year | Sequencing Method | Sample Size | Notes | Reference |
|---|---|---|---|---|---|
| Blekhman et al. | 2015 | Shotgun metagenomic | n = 93 | Variants in the LCT gene correlated with abundance of Bifidobacterium (p = 1.16 × 10−5). | [95] |
| Davenport et al. | 2015 | 16S | n = 127 | SNPs in regions of the PLD1 gene associated with abundance of genus Akkernabsia. | [96] |
| Wang et al. | 2016 | 16S | n = 182 | Forty-two loci included variants in VDR gene-encoding (vitamin D receptor) associated with beta diversity (p < 5 × 10−8). | [97] |
| Turpin et al. | 2016 | 16S | n = 1098 (discovery cohort) n = 463 (replication cohort) | Identification of 20 possibly heritable taxa. | [85,86,93] |
| Bonder et al. | 2016 | Shotgun Metagenomic | n = 1514 | Confirmation of variants in the LCT gene correlated with abundance of Bifidobacterium. Nine new human loci associated with bacterial taxa and 33 loci associated with bacterial pathways. | [98] |
| Rothschild et al. | 2018 | Shotgun Metagenomic and 16S | n = 1046 | No significant association detected. | [100] |
| Kurilshikov et al. | 2021 | 16S | n = 18340 | One SNP in the LCT locus correlated with abundance of Bifidobacterium (p = 1.28 × 10−20). FUT2 locus was suggestively associated with the abundance of Ruminococcus torques. | [88] |
| Rühlemann et al. | 2021 | 16S | n = 8956 | Thirty-eight genetic loci found to be associated with single bacteria and overall microbiome composition. ABO group was suggestively associated with the abundance of Faecalibacterium (P Meta = 6.16 × 10−9) and Bacteroides (P Meta = 3.65 × 10−10). | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maglione, A.; Zuccalà, M.; Tosi, M.; Clerico, M.; Rolla, S. Host Genetics and Gut Microbiome: Perspectives for Multiple Sclerosis. Genes 2021, 12, 1181. https://doi.org/10.3390/genes12081181
Maglione A, Zuccalà M, Tosi M, Clerico M, Rolla S. Host Genetics and Gut Microbiome: Perspectives for Multiple Sclerosis. Genes. 2021; 12(8):1181. https://doi.org/10.3390/genes12081181
Chicago/Turabian StyleMaglione, Alessandro, Miriam Zuccalà, Martina Tosi, Marinella Clerico, and Simona Rolla. 2021. "Host Genetics and Gut Microbiome: Perspectives for Multiple Sclerosis" Genes 12, no. 8: 1181. https://doi.org/10.3390/genes12081181
APA StyleMaglione, A., Zuccalà, M., Tosi, M., Clerico, M., & Rolla, S. (2021). Host Genetics and Gut Microbiome: Perspectives for Multiple Sclerosis. Genes, 12(8), 1181. https://doi.org/10.3390/genes12081181