A Causal Web between Chronotype and Metabolic Health Traits
Abstract
:1. Introduction
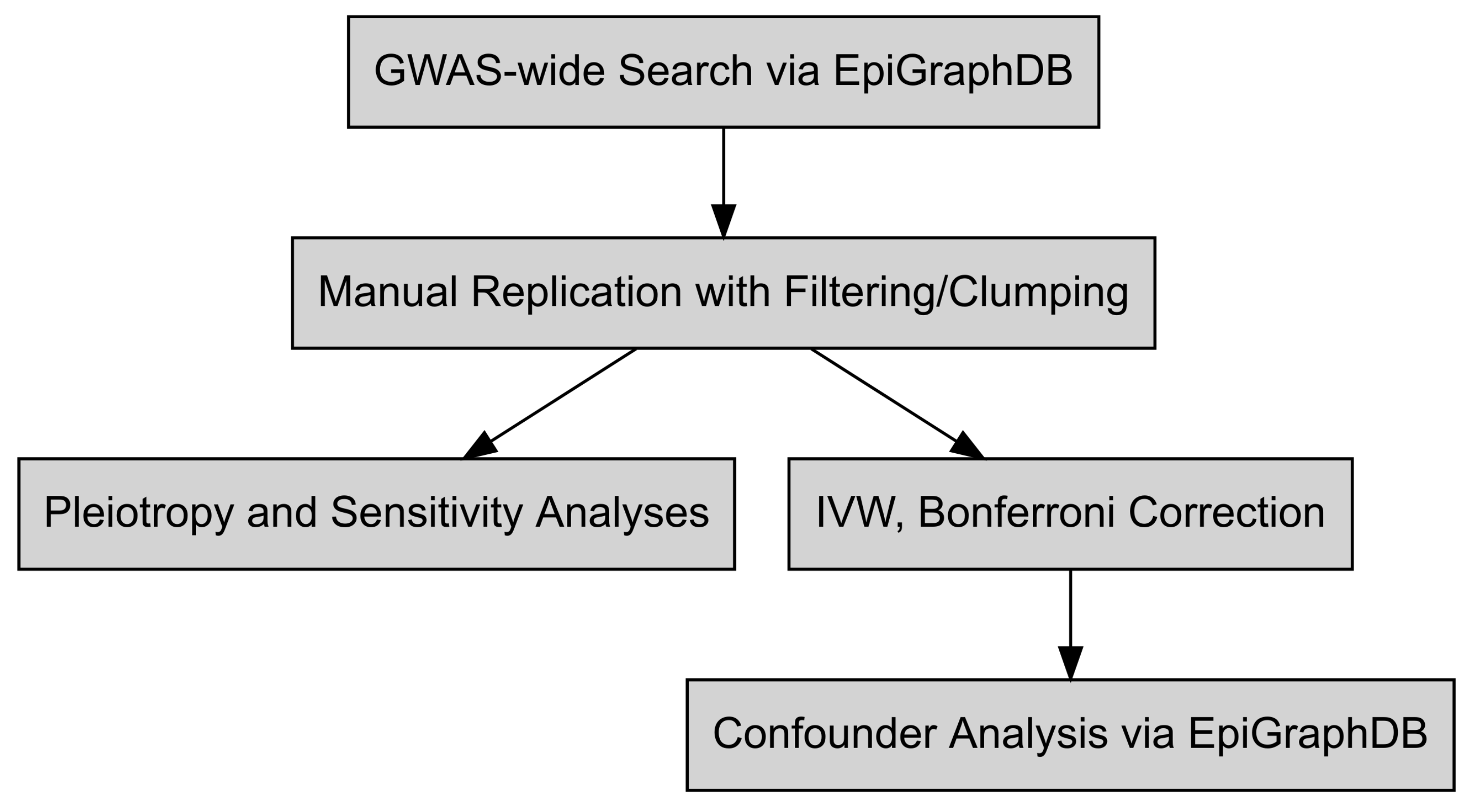
2. Materials and Methods
2.1. Mendelian Randomization Investigations
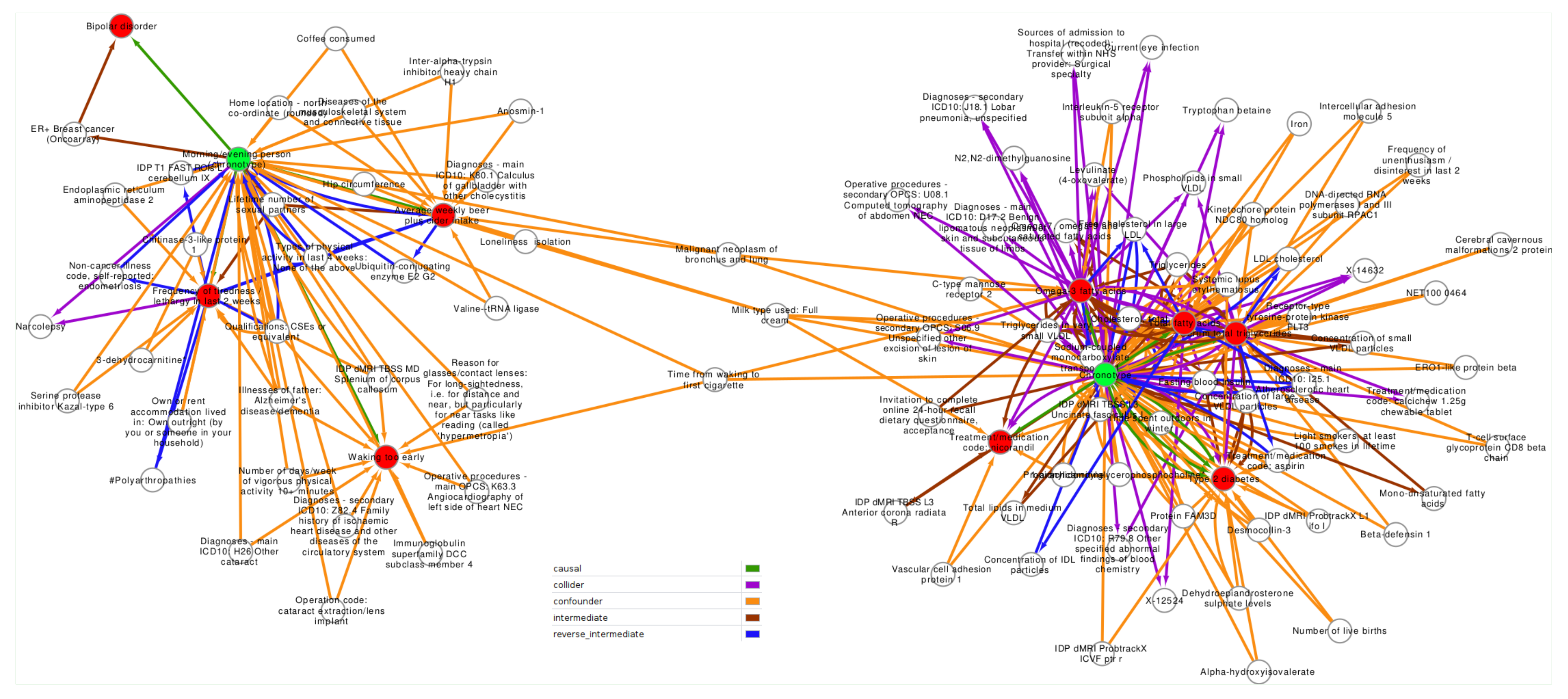
2.2. Confounder and Intermediate Analysis
3. Results
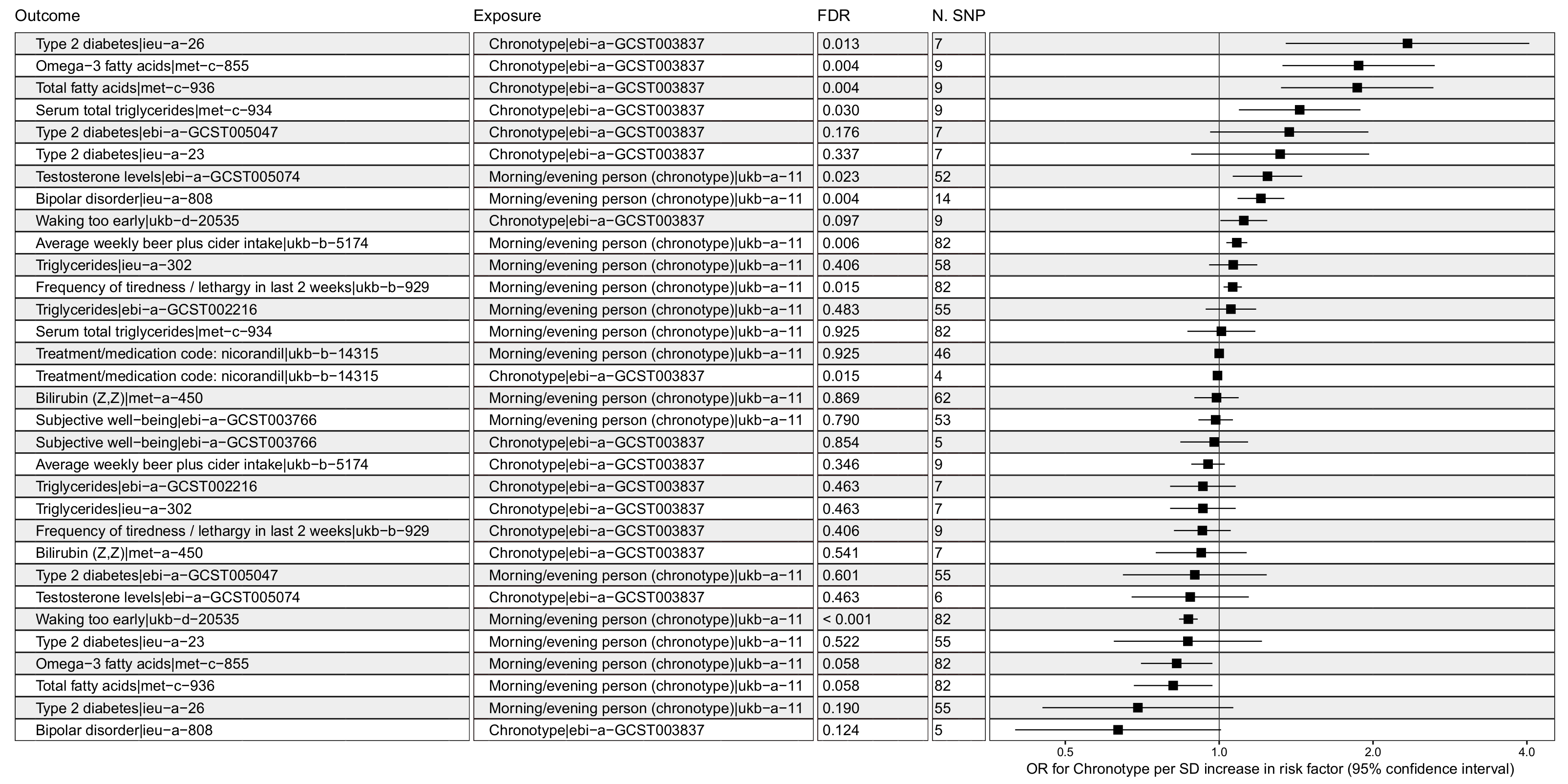
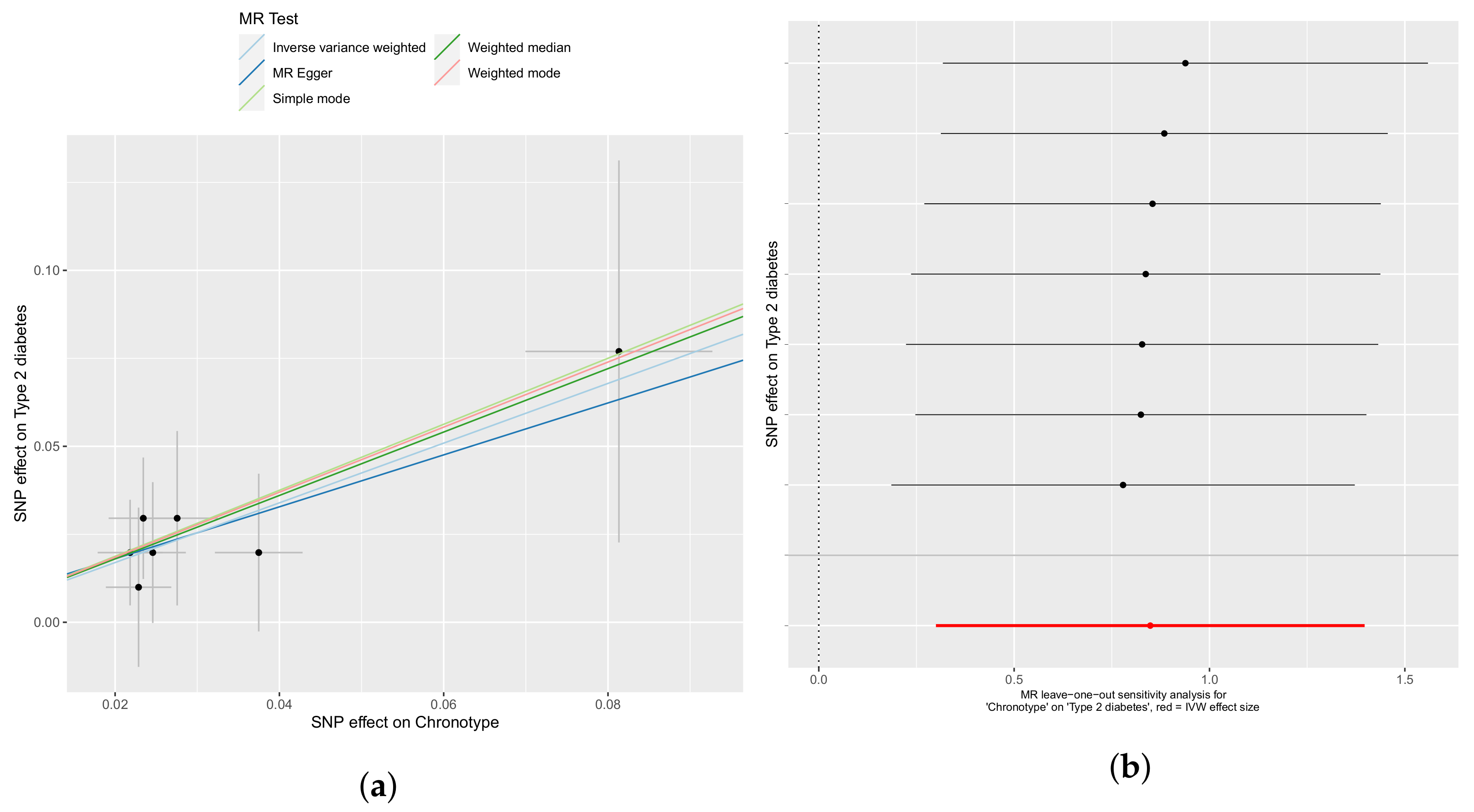
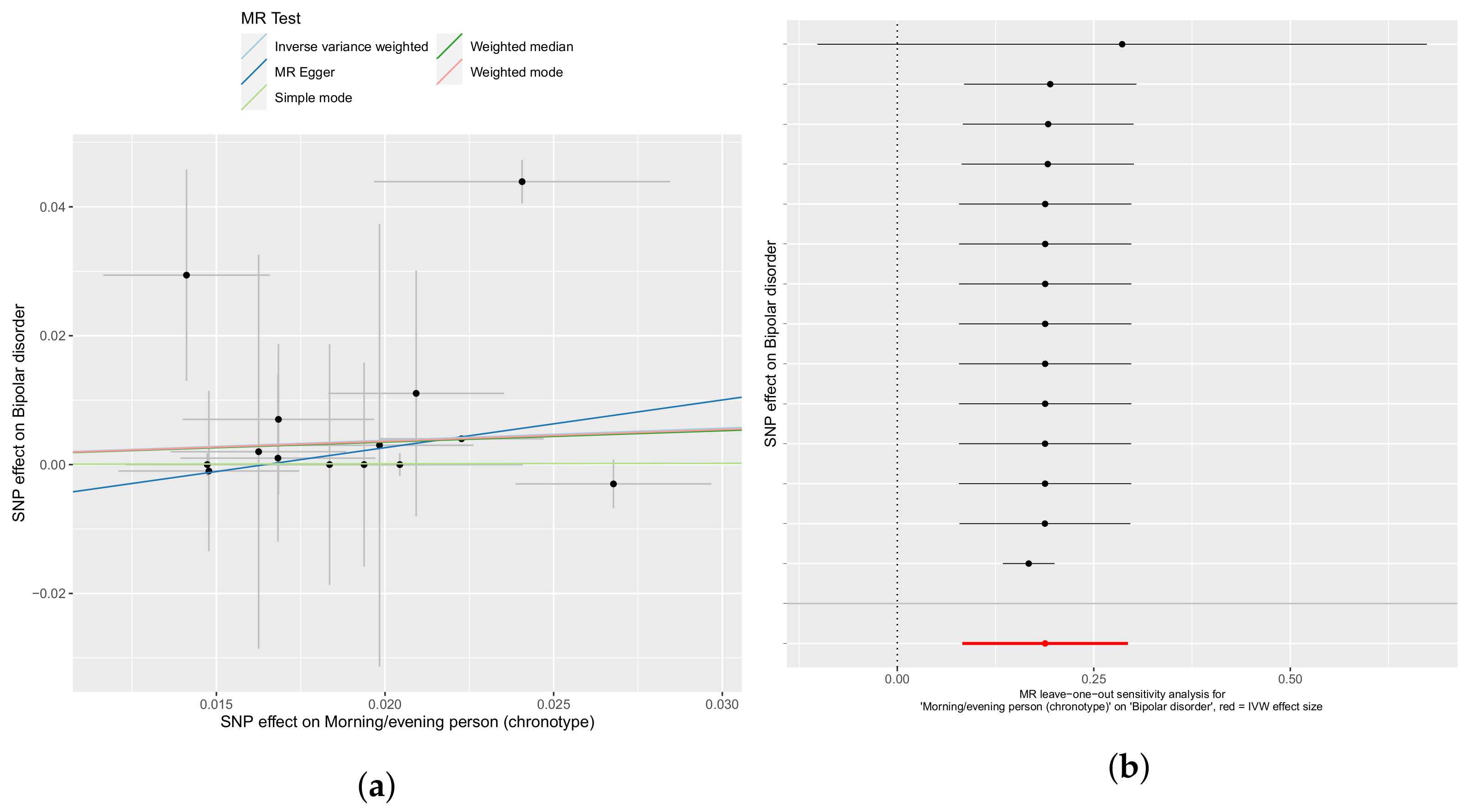
3.1. Chronotype Influences on Diabetes, Alcohol Consumption, and Bipolar Disorder
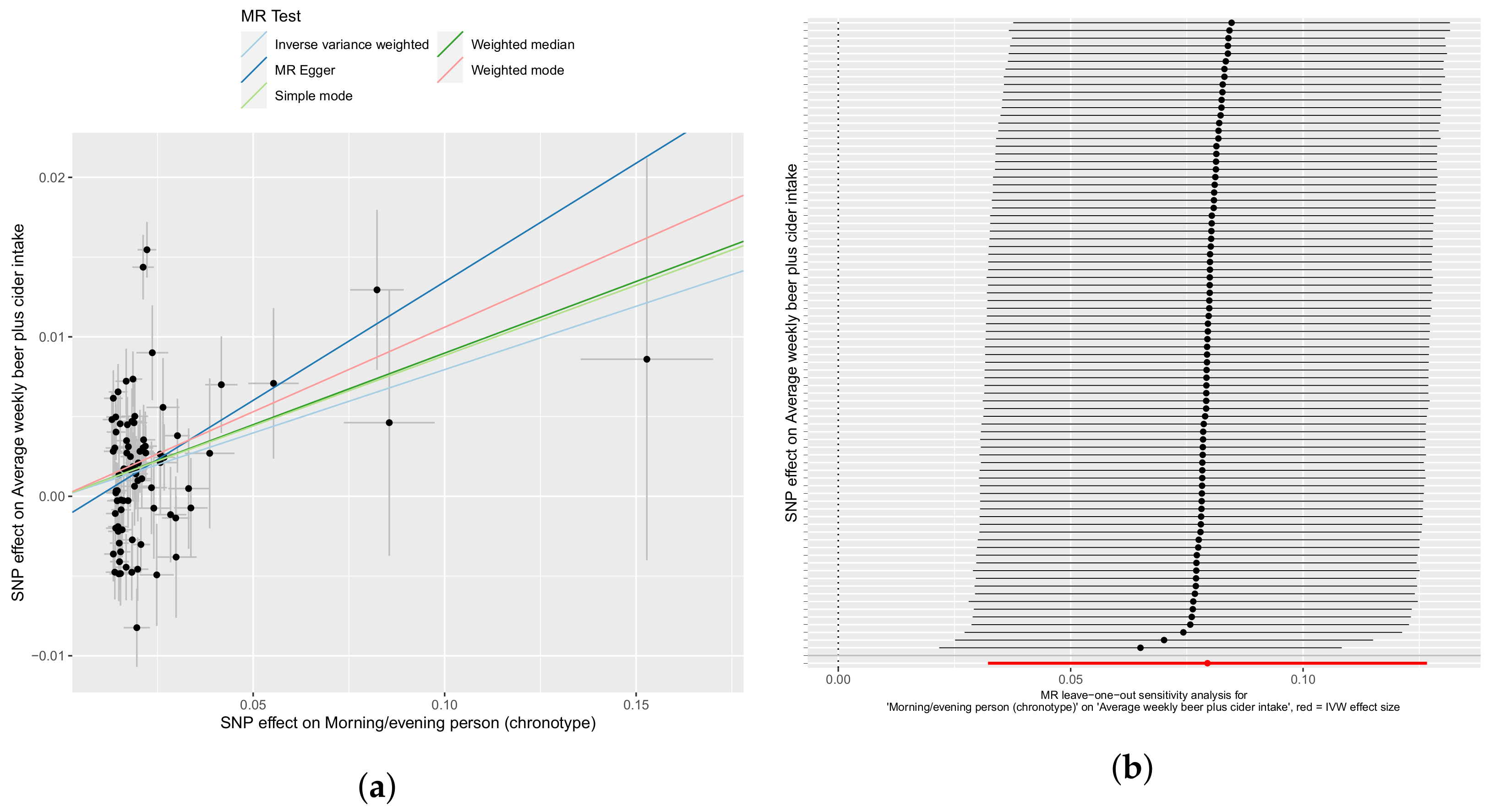
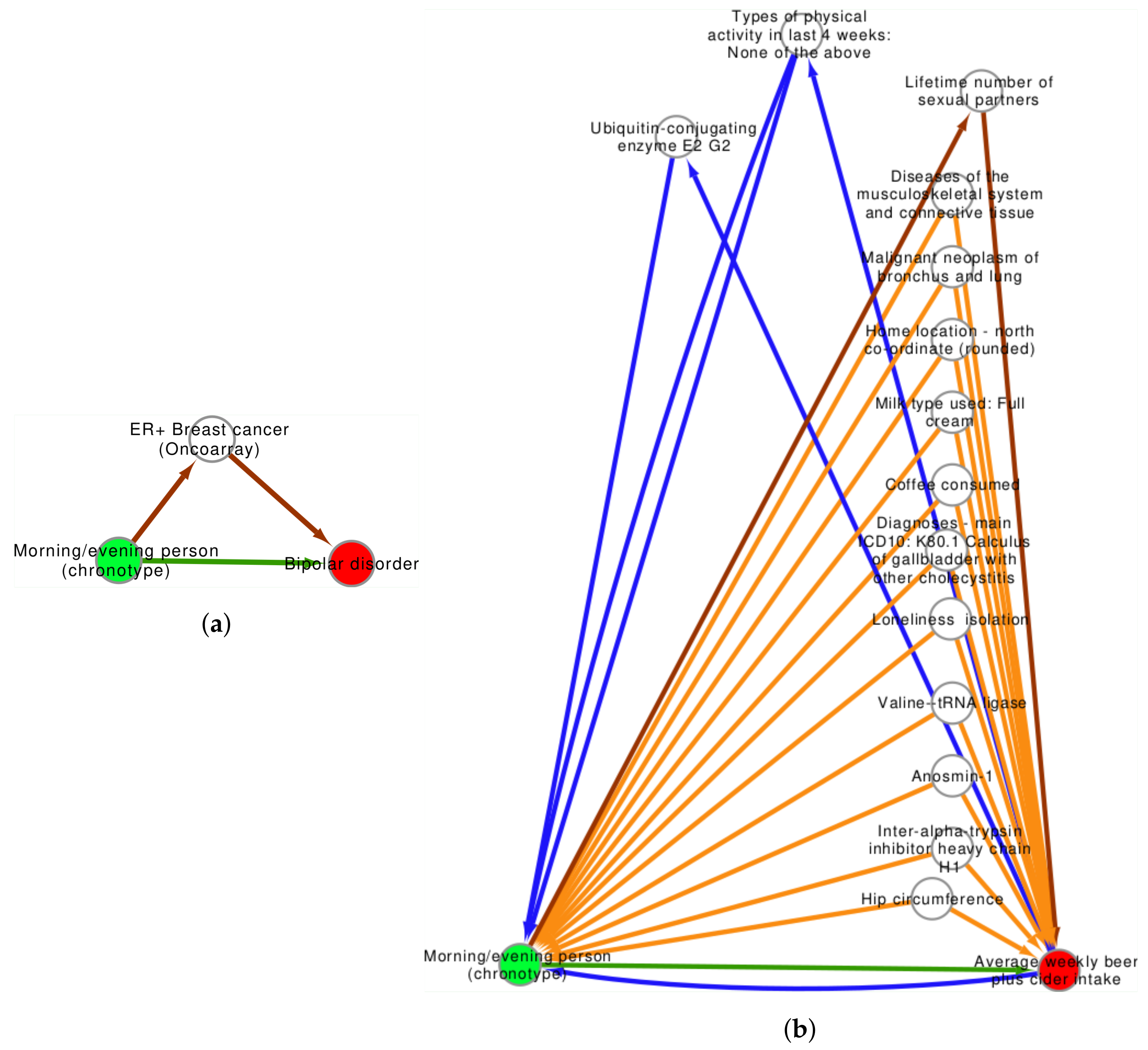
3.2. Confounder Case Studies: Bi-Polar Disorder and Alcohol Intake
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association study |
| MR | Mendelian randomization |
| IVW | Inverse-variance weighted |
| OR | Odds ratio |
| SNP | Single nucleotide polymorphism |
| LD | linkage disequilibrium |
| IV | Instrumental variable |
| MBE | mode-based estimator |
| FDR | alse discovery rate |
| T2DM | Type 2 diabetes melletus |
| ER+ | Estrogen Receptor positive |
| VLDL | Very low density lipoprotein |
References
- Lind, M.J.; Brick, L.A.; Gehrman, P.R.; Duncan, L.E.; Gelaye, B.; Maihofer, A.X.; Nievergelt, C.M.; Nugent, N.R.; Stein, M.B.; Amstadter, A.B. Psychiatric Genomics Consortium Posttraumatic Stress Disorder. Molecular genetic overlap between posttraumatic stress disorder and sleep phenotypes. Sleep 2020, 43. [Google Scholar] [CrossRef]
- Adams, C.D.; Neuhausen, S.L. Evaluating causal associations between chronotype and fatty acids and between fatty acids and type 2 diabetes: A Mendelian randomization study. Nutr. Metab. Cardiovasc. Dis. NMCD 2019, 29, 1176–1184. [Google Scholar] [CrossRef]
- Richmond, R.C.; Anderson, E.L.; Dashti, H.S.; Jones, S.E.; Lane, J.M.; Strand, L.B.; Brumpton, B.; Rutter, M.K.; Wood, A.R.; Straif, K.; et al. Investigating causal relations between sleep traits and risk of breast cancer in women: Mendelian randomisation study. BMJ (Clin. Res. Ed.) 2019, 365, l2327. [Google Scholar] [CrossRef] [Green Version]
- Gibson, M.; Munafò, M.R.; Taylor, A.E.; Treur, J.L. Evidence for Genetic Correlations and Bidirectional, Causal Effects Between Smoking and Sleep Behaviors. Nicotine Tob. Res. Off. J. Soc. Res. Nicotine Tob. 2019, 21, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Treur, J.L.; Gibson, M.; Taylor, A.E.; Rogers, P.J.; Munafò, M.R. Investigating genetic correlations and causal effects between caffeine consumption and sleep behaviours. J. Sleep Res. 2018, 27, e12695. [Google Scholar] [CrossRef] [Green Version]
- Lane, J.M.; Vlasac, I.; Anderson, S.G.; Kyle, S.D.; Dixon, W.G.; Bechtold, D.A.; Gill, S.; Little, M.A.; Luik, A.; Loudon, A.; et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat. Commun. 2016, 7, 10889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Liu, Y.; Elsworth, B.; Erola, P.; Haberland, V.; Hemani, G.; Lyon, M.; Zheng, J.; Lloyd, O.; Vabistsevits, M.; Gaunt, T.R. EpiGraphDB: A database and data mining platform for health data science. Bioinformatics 2020. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Howie, B.; McCarthy, S.; Memari, Y.; Walter, K.; Min, J.L.; Danecek, P.; Malerba, G.; Trabetti, E.; Zheng, H.F.; et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015, 6, 8111. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, D.; Patel, C.J. Conducting a Reproducible Mendelian Randomization Analysis Using the R Analytic Statistical Environment. Curr. Protoc. Hum. Genet. 2019, 101, e82. [Google Scholar] [CrossRef]
- Benyamin, B.; Visscher, P.M.; McRae, A.F. Family-based genome-wide association studies. Pharmacogenomics 2009, 10, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.; Green, S. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions, 5.1.0 [updated march 2011] ed.; The Coochrane Collaboration: 2011. Available online: www.training.cochrane.org/handbook (accessed on 5 November 2012).
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Tilling, K.; Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017, 13, e1007081. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ma, J.; Yao, K.; Su, W.; Tan, B.; Wu, X.; Huang, X.; Li, T.; Yin, Y.; Tosini, G.; et al. Circadian Rhythms and Obesity: Timekeeping Governs Lipid Metabolism. J. Pineal Res. 2020, e12682. [Google Scholar] [CrossRef]
- Socaciu, A.I.; Ionuţ, R.; Socaciu, M.A.; Ungur, A.P.; Bârsan, M.; Chiorean, A.; Socaciu, C.; Râjnoveanu, A.G. Melatonin, an ubiquitous metabolic regulator: Functions, mechanisms and effects on circadian disruption and degenerative diseases. Rev. Endocr. Metab. Disord. 2020. [Google Scholar] [CrossRef]
- Pan, X.; Mota, S.; Zhang, B. Circadian Clock Regulation on Lipid Metabolism and Metabolic Diseases. Adv. Exp. Med. Biol. 2020, 1276, 53–66. [Google Scholar] [CrossRef]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef] [Green Version]
- Gantenbein, M.; Attolini, L.; Bruguerolle, B. Nicorandil affects diurnal rhythms of body temperature, heart rate and locomotor activity in rats. Eur. J. Pharmacol. 1998, 346, 125–130. [Google Scholar] [CrossRef]
- Kusama, Y.; Kodani, E.; Nakagomi, A.; Otsuka, T.; Atarashi, H.; Kishida, H.; Mizuno, K. Variant angina and coronary artery spasm: The clinical spectrum, pathophysiology, and management. J. Nippon. Med. Sch. Nippon Ika Daigaku Zasshi 2011, 78, 4–12. [Google Scholar] [CrossRef] [Green Version]
- Hisler, G.C.; Rothenberger, S.D.; Clark, D.B.; Hasler, B.P. Is there a 24-hour rhythm in alcohol craving and does it vary by sleep/circadian timing? Chronobiol. Int. 2021, 38, 109–121. [Google Scholar] [CrossRef]
- Ferrer, A.; Costas, J.; Gratacos, M.; Martínez-Amorós, È.; Labad, J.; Soriano-Mas, C.; Palao, D.; Menchón, J.M.; Crespo, J.M.; Urretavizcaya, M.; et al. Clock gene polygenic risk score and seasonality in major depressive disorder and bipolar disorder. Genes Brain Behav. 2020, 19, e12683. [Google Scholar] [CrossRef]
- Romo-Nava, F.; Blom, T.J.; Cuellar-Barboza, A.B.; Winham, S.J.; Colby, C.L.; Nunez, N.A.; Biernacka, J.M.; Frye, M.A.; McElroy, S.L. Evening chronotype as a discrete clinical subphenotype in bipolar disorder. J. Affect. Disord. 2020, 266, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, J.M. Thinking Clearly About Correlations and Causation: Graphical Causal Models for Observational Data. Adv. Methods Pract. Psychol. Sci. 2018, 1, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Allen, N.E.; Sudlow, C.; Peakman, T.; Collins, R.; Biobank, O.b.O.U. UK Biobank Data: Come and Get It. Sci. Transl. Med. 2014, 6, 224ed4. [Google Scholar] [CrossRef]
- Labrecque, J.; Swanson, S.A. Understanding the Assumptions Underlying Instrumental Variable Analyses: A Brief Review of Falsification Strategies and Related Tools. Curr. Epidemiol. Rep. 2018, 5, 214–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, F.; Acharjee, A.; Williams, J.A.; Russ, D.; Bravo-Merodio, L.; Gkoutos, G.V. Biomarker prioritisation and power estimation using ensemble gene regulatory network inference. Int. J. Mol. Sci. 2020, 21, 7886. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, J.A.; Russ, D.; Bravo-Merodio, L.; Cardoso, V.R.; Pendleton, S.C.; Aziz, F.; Acharjee, A.; Gkoutos, G.V. A Causal Web between Chronotype and Metabolic Health Traits. Genes 2021, 12, 1029. https://doi.org/10.3390/genes12071029
Williams JA, Russ D, Bravo-Merodio L, Cardoso VR, Pendleton SC, Aziz F, Acharjee A, Gkoutos GV. A Causal Web between Chronotype and Metabolic Health Traits. Genes. 2021; 12(7):1029. https://doi.org/10.3390/genes12071029
Chicago/Turabian StyleWilliams, John A., Dominic Russ, Laura Bravo-Merodio, Victor Roth Cardoso, Samantha C. Pendleton, Furqan Aziz, Animesh Acharjee, and Georgios V. Gkoutos. 2021. "A Causal Web between Chronotype and Metabolic Health Traits" Genes 12, no. 7: 1029. https://doi.org/10.3390/genes12071029
APA StyleWilliams, J. A., Russ, D., Bravo-Merodio, L., Cardoso, V. R., Pendleton, S. C., Aziz, F., Acharjee, A., & Gkoutos, G. V. (2021). A Causal Web between Chronotype and Metabolic Health Traits. Genes, 12(7), 1029. https://doi.org/10.3390/genes12071029








