Genome-Wide Association Study Reveals a Genomic Region Associated with Mite-Recruitment Phenotypes in the Domesticated Grapevine (Vitis vinifera)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Association Panel
2.2. Phenotyping
2.3. Genome-wide Association Study Analysis (GWAS) and Genomic Prediction
3. Results
3.1. Phenotyping
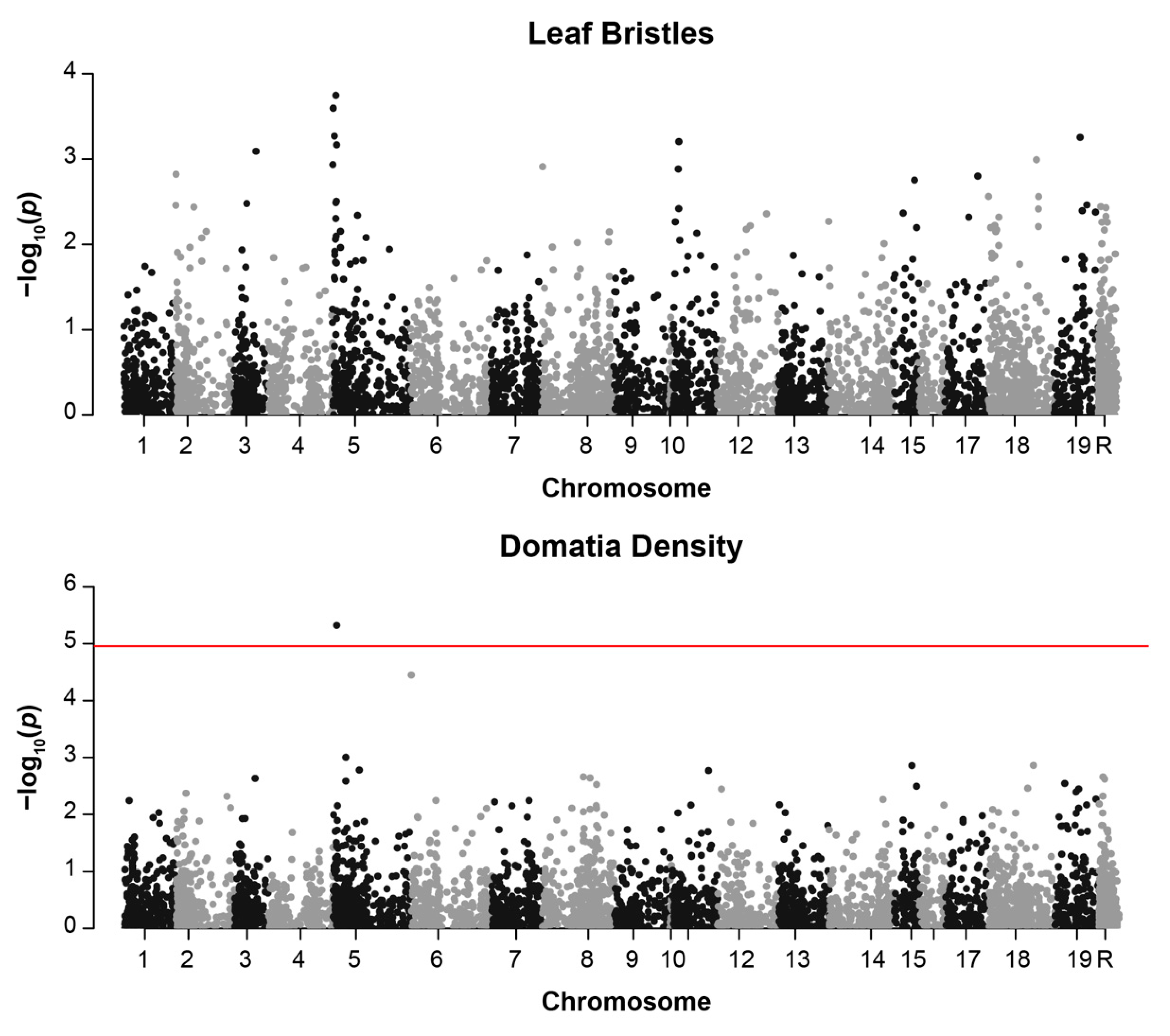
3.2. GWAS Results
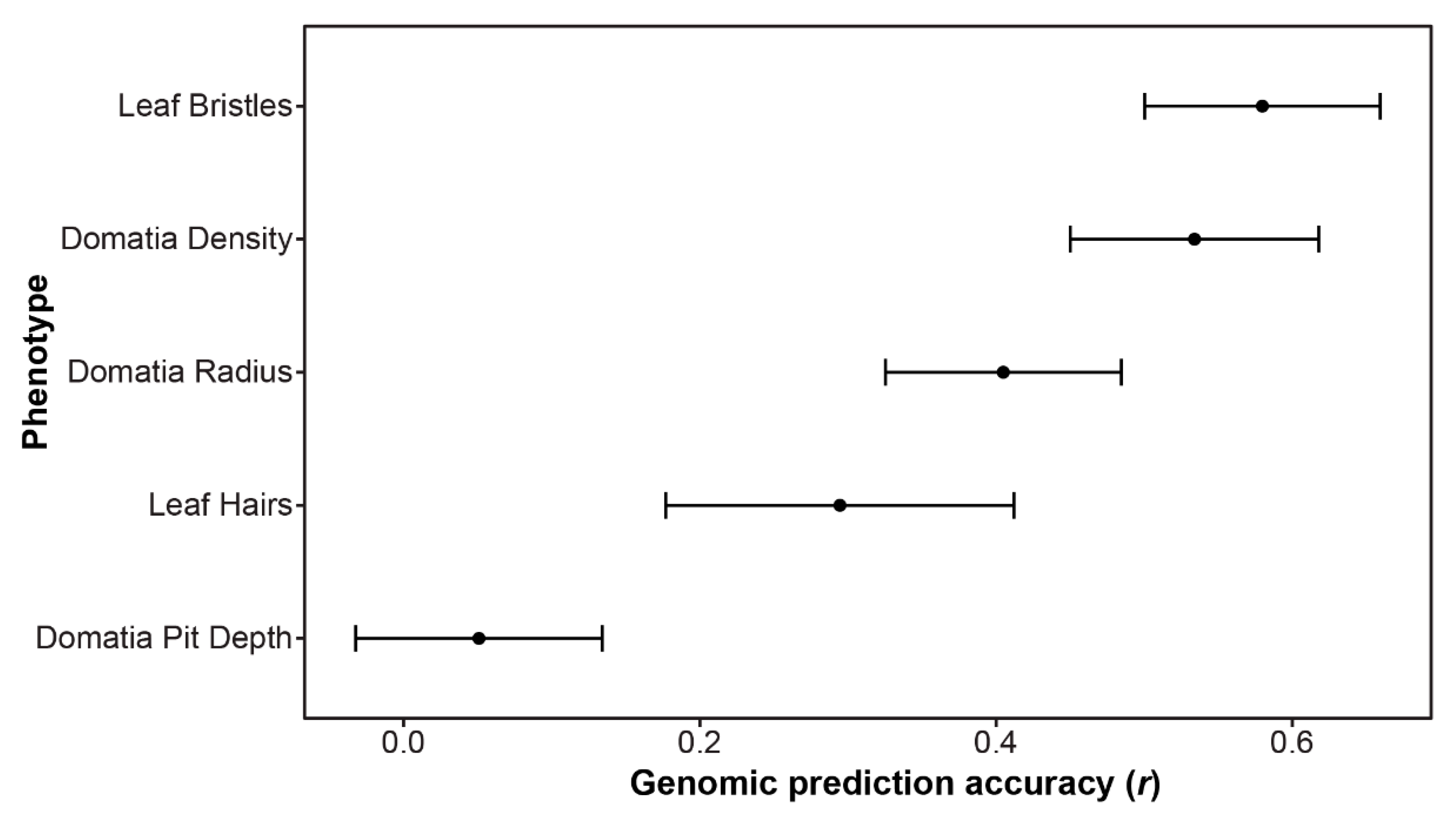
3.3. Genomic Prediction Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heil, M. Indirect defence via tritrophic interactions. New Phytol. 2008, 178, 41–61. [Google Scholar] [CrossRef]
- Bronstein, J.L.; Alarcón, R.; Geber, M. The evolution of plant–insect mutualisms. New Phytol. 2006, 172, 412–428. [Google Scholar] [CrossRef]
- Pearse, I.S.; Lopresti, E.; Schaeffer, R.N.; Wetzel, W.C.; Mooney, K.A.; Ali, J.G.; Ode, P.J.; Eubanks, M.D.; Bronstein, J.L.; Weber, M.G. Generalising indirect defence and resistance of plants. Ecol. Lett. 2020, 23, 1137–1152. [Google Scholar] [CrossRef]
- Lundstrom, A.N. Lundström Pflanzenbiologische Studien II. Die Anpassungen Der Pflanzen an Thiere, I. Von Domatia. Nova Acta Regiae Soc. Sci. 1887, 13, 1–87. [Google Scholar]
- O’Dowd, D.J.; Willson, M.F. Associations between mites and leaf domatia. Trends Ecol. Evol. 1991, 6, 179–182. [Google Scholar] [CrossRef]
- O’Dowd, D.J.; Willson, M.F. Leaf domatia and mites on Australasian plants: Ecological and evolutionary implications. Biol. J. Linn. Soc. 1989, 37, 191–236. [Google Scholar] [CrossRef]
- English-Loeb, G.; Norton, A.P.; Walker, M.A. Behavioral and Population Consequences of Acarodomatia in Grapes on Phy-toseiid Mites (Mesostigmata) and Implications for Plant Breeding. Entomol. Exp. Appl. 2002, 104, 307–319. [Google Scholar] [CrossRef]
- Nishida, S.; Naiki, A.; Nishida, T. Morphological variation in leaf domatia enables coexistence of antagonistic mites in Cinnamomum camphora. Can. J. Bot. 2005, 83, 93–101. [Google Scholar] [CrossRef]
- Norton, A.P.; English-Loeb, G.; Gadoury, D.; Seem, R.C. Mycophagous Mites and Foliar Pathogens: Leaf Domatia Mediate Tritrophic Interactions in Grapes. Ecology 2000, 81, 490–499. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Karban, R. Domatia mediate plantarthropod mutualism. Nat. Cell Biol. 1997, 387, 562–563. [Google Scholar] [CrossRef]
- English-Loeb, G.; Norton, A.P.; Gadoury, D.; Seem, R.; Wilcox, W. Tri-Trophic Interactions among Grapevines, a Fungal Pathogen, and a Mycophagous Mite. Ecol. Appl. 2005, 15, 1679–1688. [Google Scholar] [CrossRef]
- Walter, D.E. Living on Leaves: Mites, Tomenta, and Leaf Domatia. Annu. Rev. Entomol. 1996, 41, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Karban, R.; English-Loeb, G.; Walker, M.A.; Thaler, J. Abundance of phytoseiid mites on Vitis species: Effects of leaf hairs, domatia, prey abundance and plant phylogeny. Exp. Appl. Acarol. 1995, 19, 189–197. [Google Scholar] [CrossRef]
- Grostal, P.; O’Dowd, D.J. Plants, Mites and Mutualism—Leaf Domatia and the Abundance and Reproduction of Mites on Viburnum Tinus (Caprifoliaceae). Oecologia 1994, 97, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.A. Leaf structures affect predatory mites (Acari: Phytoseiidae) and biological control: A review. Exp. Appl. Acarol. 2013, 62, 1–17. [Google Scholar] [CrossRef]
- Loughner, R.; Goldman, K.; Loeb, G.; Nyrop, J. Influence of Leaf Trichomes on Predatory Mite (Typhlodromus Pyri) Abun-dance in Grape Varieties. Exp. Appl. Acarol. 2008, 45, 111–122. [Google Scholar] [CrossRef]
- Hanley, M.E.; Lamont, B.B.; Fairbanks, M.M.; Rafferty, C.M. Plant structural traits and their role in anti-herbivore defence. Perspect. Plant Ecol. Evol. Syst. 2007, 8, 157–178. [Google Scholar] [CrossRef]
- Werker, E. Trichome diversity and development. In Advances in Botanical Research; Elsevier BV: Amsterdam, The Netherlands, 2000; Volume 31, pp. 1–35. [Google Scholar]
- Barba, P.; Loughner, R.; Wentworth, K.; Nyrop, J.P.; Loeb, G.M.; Reisch, B.I. A QTL associated with leaf trichome traits has a major influence on the abundance of the predatory mite Typhlodromus pyri in a hybrid grapevine population. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- English-Loeb, G.; Norton, A.P.; Gadoury, D.; Seem, R.; Wilcox, W. Biological Control of Grape Powdery Mildew Using My-cophagous Mites. Plant Dis. 2007, 91, 421–429. [Google Scholar] [CrossRef]
- Prischmann, D.A.; Croft, B.A.; Luh, H.-K. Biological Control of Spider Mites on Grape by Phytoseiid Mites (Acari: Tetranychidae, Phytoseiidae): Emphasis on Regional Aspects. J. Econ. Èntomol. 2002, 95, 340–347. [Google Scholar] [CrossRef]
- Duso, C.; Pozzebon, A.; Capuzzo, C.; Malagnini, V.; Otto, S.; Borgo, M. Grape downy mildew spread and mite seasonal abundance in vineyards: Effects on Tydeus caudatus and its predators. Biol. Control. 2005, 32, 143–154. [Google Scholar] [CrossRef]
- Bois, B.; Zito, S.; Calonnec, A. Climate vs grapevine pests and diseases worldwide: The first results of a global survey. OENO One 2017, 51, 133–139. [Google Scholar] [CrossRef]
- MKF. Research Impact of Wine, Grapes, and Grape Products on the American Economy; MKF Res. Bull.; MKF: St. Helena, CA, USA, 2007. [Google Scholar]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D. Ge-netic Structure and Domestication History of the Grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; Duan, S.; Sheng, J.; Zhu, S.; Ni, X.; Shao, J.; Liu, C.; Nick, P.; Du, F.; Fan, P.; et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Massonnet, M.; Sanjak, J.S.; Cantu, D.; Gaut, B.S. Evolutionary Genomics of Grape (Vitis vinifera Ssp. Vinifera) Do-mestication. Proc. Natl. Acad. Sci. USA 2017, 114, 11715–11720. [Google Scholar] [CrossRef] [Green Version]
- Migicovsky, Z.; Sawler, J.; Gardner, K.M.; Aradhya, M.K.; Prins, B.H.; Schwaninger, H.R.; Bustamante, C.D.; Buckler, E.S.; Zhong, G.-Y.; Brown, P.J.; et al. Patterns of genomic and phenomic diversity in wine and table grapes. Hortic. Res. 2017, 4, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Myles, S.; Chia, J.-M.; Hurwitz, B.; Simon, C.; Zhong, G.Y.; Buckler, E.; Ware, D. Rapid Genomic Characterization of the Ge-nus Vitis. PLoS ONE 2010, 5, e8219. [Google Scholar] [CrossRef] [Green Version]
- Money, D.; Gardner, K.; Migicovsky, Z.; Schwaninger, H.; Zhong, G.-Y.; Myles, S. LinkImpute: Fast and Accurate Genotype Imputation for Nonmodel Organisms. G3 Genes Genomes Genet. 2015, 5, 2383–2390. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Map-ping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Galet, P. A Practical Ampelography (Grapevine Identification) Luicie T; NCROL: London, UK, 1979. [Google Scholar]
- Ipgri, U.; OIV. Descriptors for Grapevine (Vitis Spp.). International Union for the Protection of New Varieties of Plants, Geneva, Switzerland/Office International de La Vigne et Du Vin, Paris, France/International Plant Genetic Resources Institute, Rome, Italy. This Publ. 1997. Available online: http://www.cgiar.org/ipgri/IPGRI UPOV OIV Via delle Sette Chiese 142 4 (accessed on 2 May 2021).
- Ma, Z.; Wen, J.; Ickert-Bond, S.M.; Chen, L.-Q.; Liu, X.-Q. Morphology, Structure, and Ontogeny of Trichomes of the Grape Genus (Vitis, Vitaceae). Front. Plant Sci. 2016, 7, 704. [Google Scholar] [CrossRef] [Green Version]
- Gago, P.; Conéjéro, G.; Martínez, M.; Boso, S.; This, P.; Verdeil, J.-L. Microanatomy of leaf trichomes: Opportunities for improved ampelographic discrimination of grapevine (Vitis vinifera L.) cultivars. Aust. J. Grape Wine Res. 2016, 22, 494–503. [Google Scholar] [CrossRef]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Revelle, W.; Revelle, M.W. Package ‘Psych’. Compr. R Arch. Netw. 2015, 337, 338. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Package ‘Corrplot’. Statistician 2017, 56, e24. [Google Scholar]
- Schloerke, B.; Crowley, J.; Cook, D.; Briatte, F.; Marbach, M.; Thoen, E.; Elberg, A.; Larmarange, J. Ggally: Extension to Ggplot2; R Package Version 2018; R Core Team: Vienna, Austria, 2018; Volume 1. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Com-puting: Vienna, Austria, 2020. [Google Scholar]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Canaguier, A.; Grimplet, J.; di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom. Data 2017, 14, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Tiede, T.; Mohammadi, M.; Smith, K.P. PopVar: Genomic Breeding Tools: Genetic Variance Prediction and Cross-Validation. R Package Version 121. 2015. Available online: https://CRAN.R-project.org/package=PopVar (accessed on 2 May 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysi; Springer: New York, NY, USA, 2016. [Google Scholar]
- Zhang, Z.; Ren, C.; Zou, L.; Wang, Y.; Li, S.; Liang, Z. Characterization of the GATA gene family in Vitis vinifera: Genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome 2018, 61, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Peng, J.; Yin, X.; Li, M.; Xiang, G.; Wang, Y.; Lei, Y.; Xu, Y. Importin-αs are required for the nuclear localization and function of the Plasmopara viticola effector PvAVH53. Hortic. Res. 2021, 8, 1–12. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef]
- Gan, Y.; Liu, C.; Yu, H.; Broun, P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 2007, 134, 2073–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitwood, D.H.; Ranjan, A.; Martinez, C.C.; Headland, L.R.; Thiem, T.; Kumar, R.; Covington, M.F.; Hatcher, T.; Naylor, D.T.; Zimmerman, S.; et al. A Modern Ampelography: A Genetic Basis for Leaf Shape and Venation Patterning in Grape. Plant Physiol. 2014, 164, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Behringer, C.; Bastakis, E.; Ranftl, Q.L.; Mayer, K.; Schwechheimer, C. Functional Diversification within the Family of B-GATA Transcription Factors through the Leucine-Leucine-Methionine Domain. Plant Physiol. 2014, 166, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranftl, Q.L.; Bastakis, E.; Klermund, C.; Schwechheimer, C. LLM-Domain Containing B-GATA Factors Control Different Aspects of Cytokinin-Regulated Development in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2295–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andre, H. Duges caudatus is a Tenuipalpidae and not a Tydeidae (Acari). Acarologia 2011, 51, 69–85. [Google Scholar] [CrossRef] [Green Version]
- McClure, K.A.; Gong, Y.; Song, J.; Vinqvist-Tymchuk, M.; Palmer, L.C.; Fan, L.; Burgher-Maclellan, K.; Zhang, Z.; Celton, J.-M.; Forney, C.F.; et al. Genome-wide association studies in apple reveal loci of large effect controlling apple polyphenols. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Migicovsky, Z.; Gardner, K.M.; Money, D.; Sawler, J.; Bloom, J.S.; Moffett, P.; Chao, C.T.; Schwaninger, H.; Fazio, G.; Zhong, G.; et al. Genome to Phenome Mapping in Apple Using Historical Data. Plant Genome 2016, 9, 2015-11. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Fishbein, M. Plant Defense Syndromes. Ecology 2006, 87, S132–S149. [Google Scholar] [CrossRef]
- Agrawal, A.A. A scale-dependent framework for trade-offs, syndromes, and specialization in organismal biology. Ecology 2020, 101, e02924. [Google Scholar] [CrossRef]
- Brouwer, Y.; Clifford, H. An Annotated List of Domatia-Bearing Species. Notes Jodrell Lab. 1990, 12, 1–33. [Google Scholar]
- Kono, A.; Ban, Y.; Mitani, N.; Fujii, H.; Sato, S.; Suzaki, K.; Azuma, A.; Onoue, N.; Sato, A. Development of SSR markers linked to QTL reducing leaf hair density and grapevine downy mildew resistance in Vitis vinifera. Mol. Breed. 2018, 38, 1–19. [Google Scholar] [CrossRef]
- Yin, L.; Karn, A.; Cadle-Davidson, L.; Zou, C.; Underhill, A.; Atkins, P.; Treiber, E.; Voytas, D.; Clark, M. Fine Mapping of Leaf Trichome Density Revealed a 747-kb Region on Chromosome 1 in Cold-Hardy Hybrid Wine Grape Populations. Front. Plant Sci. 2021, 12, 150. [Google Scholar] [CrossRef] [PubMed]
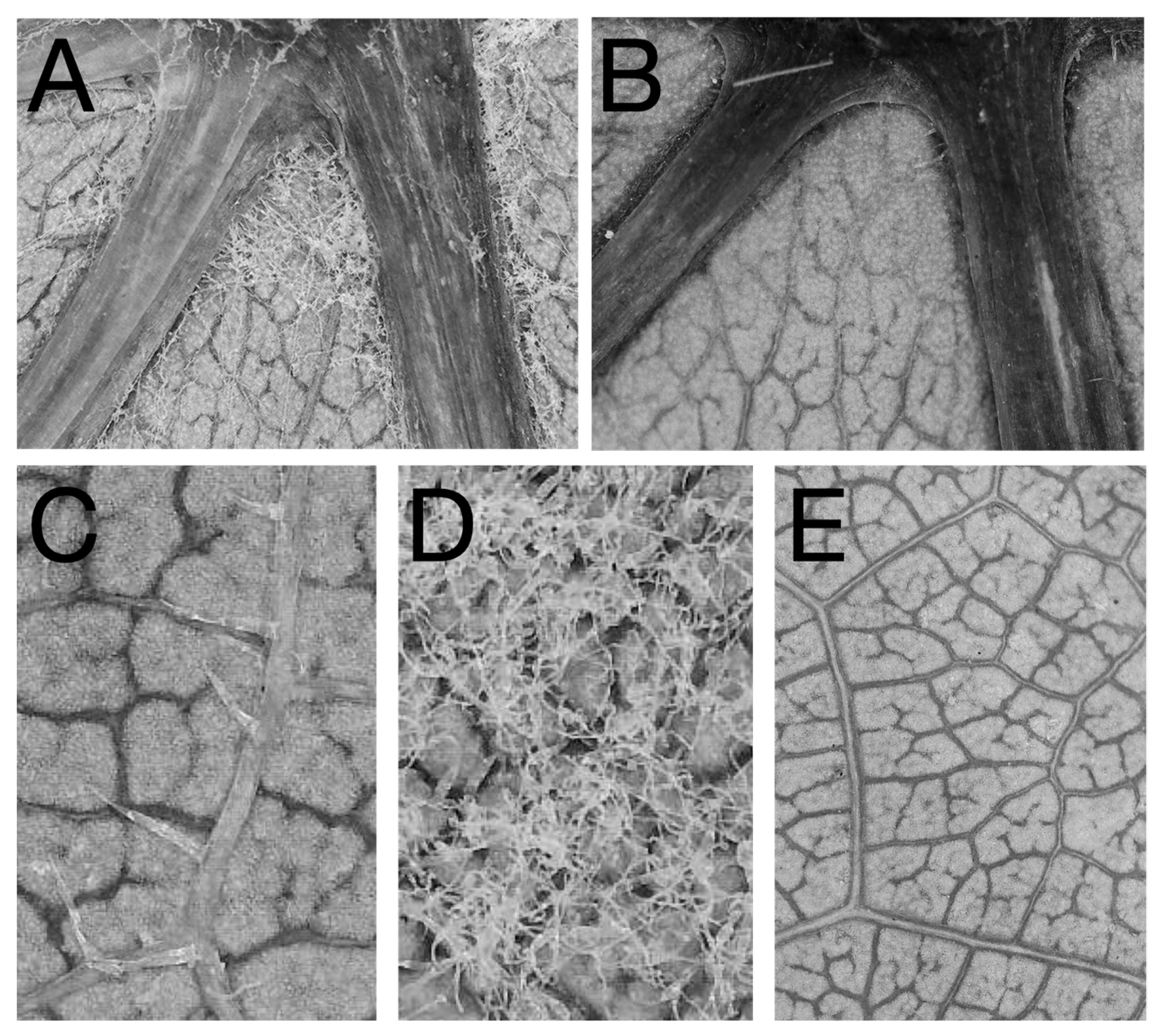
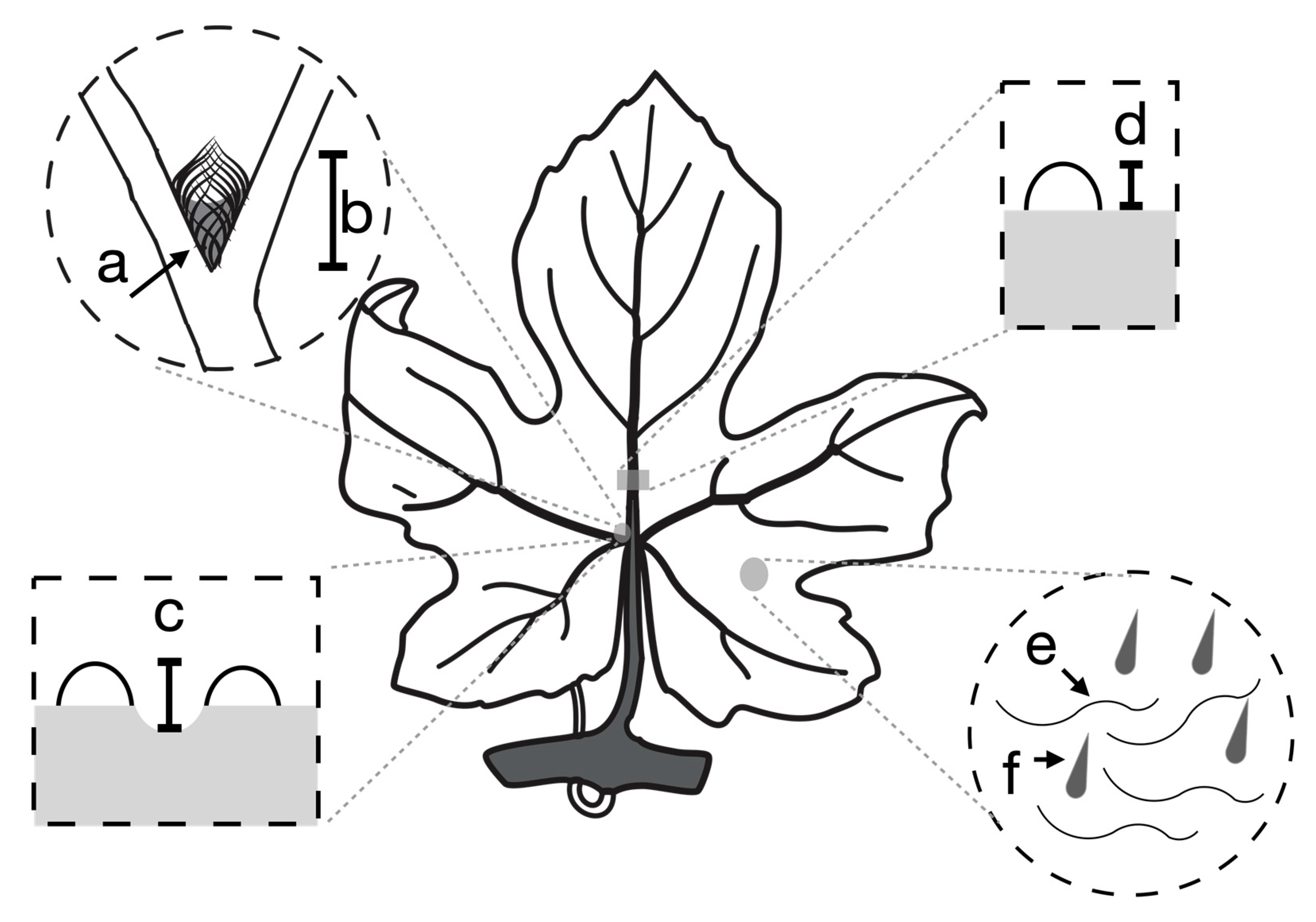

| Trait | Description | Citations Demonstrating Trait Enhances Beneficial Mite Abundance |
|---|---|---|
| Domatia size | The size of the most basal domatium just left of the midvein | [7,11] |
| Domatia density | The density of trichomes making up the most basal domatium just left of the midvein | [7,13] |
| Domatia pit depth | The degree to which the leaf lamina is depressed inside the domatia, creating a cavern for mites to occupy | [8] |
| Leaf bristles | The presence or absence of erect trichomes on the abaxial leaf lamina | [16,17,19,22] |
| Leaf hairs | The presence or absence of prostrate, ribbon-like trichomes on the abaxial leaf lamina | [16,17,19,22] |
| Gene ID v.2 | Annotation | Coordinates | Gene ID v.0 | Arabidopsis Homolog |
|---|---|---|---|---|
| VIT_205s0077g01340 | Powdery Mildew Resistance 5 | 1085706-1087504 | GSVIVG01035039001 | At5g49340 |
| VIT_205s0077g01390 | Glabrous Inflorescence 2 | 1128866-1129489 | Not present | At1g67030 |
| VIT_205s0077g01440 | Importin Alpha Isoform 1 | 1148995-1153335 | GSVIVG01035047001 | At3g06720 |
| SNP Chr5:1160194 | 1160194 | |||
| VIT_205s0077g01450 | GATA Transcription Factor 8 | 1163455-1164868 | GSVIVG01035048001 | At3g06740 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
LaPlante, E.R.; Fleming, M.B.; Migicovsky, Z.; Weber, M.G. Genome-Wide Association Study Reveals a Genomic Region Associated with Mite-Recruitment Phenotypes in the Domesticated Grapevine (Vitis vinifera). Genes 2021, 12, 1013. https://doi.org/10.3390/genes12071013
LaPlante ER, Fleming MB, Migicovsky Z, Weber MG. Genome-Wide Association Study Reveals a Genomic Region Associated with Mite-Recruitment Phenotypes in the Domesticated Grapevine (Vitis vinifera). Genes. 2021; 12(7):1013. https://doi.org/10.3390/genes12071013
Chicago/Turabian StyleLaPlante, Erika R., Margaret B. Fleming, Zoë Migicovsky, and Marjorie Gail Weber. 2021. "Genome-Wide Association Study Reveals a Genomic Region Associated with Mite-Recruitment Phenotypes in the Domesticated Grapevine (Vitis vinifera)" Genes 12, no. 7: 1013. https://doi.org/10.3390/genes12071013
APA StyleLaPlante, E. R., Fleming, M. B., Migicovsky, Z., & Weber, M. G. (2021). Genome-Wide Association Study Reveals a Genomic Region Associated with Mite-Recruitment Phenotypes in the Domesticated Grapevine (Vitis vinifera). Genes, 12(7), 1013. https://doi.org/10.3390/genes12071013






