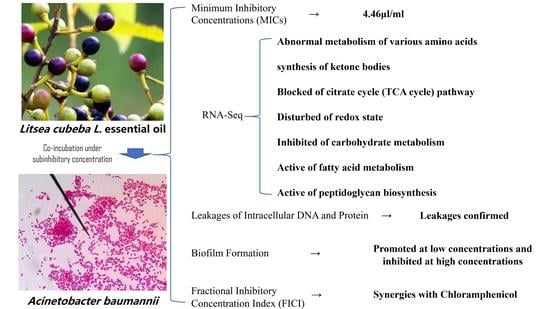
Transcriptomic Analysis of Drug-Resistance Acinetobacter baumannii under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Bacterial Strains
2.2. Determination of Minimum Inhibitory Concentration (MIC)
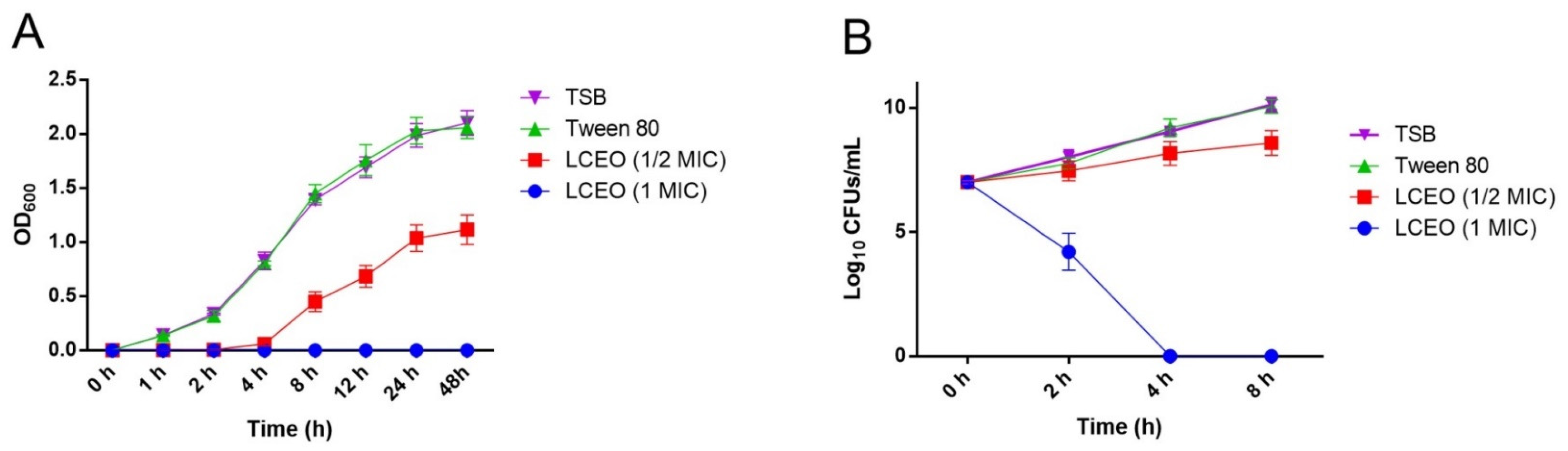
2.3. Growth Curve
2.4. Time Kill Curve
2.5. Preparation of Bacterial Samples for RNA-Seq
2.6. RNA Extraction and Purification, cDNA Library Construction for RNA-Seq, RNA-Seq and Data Analysis
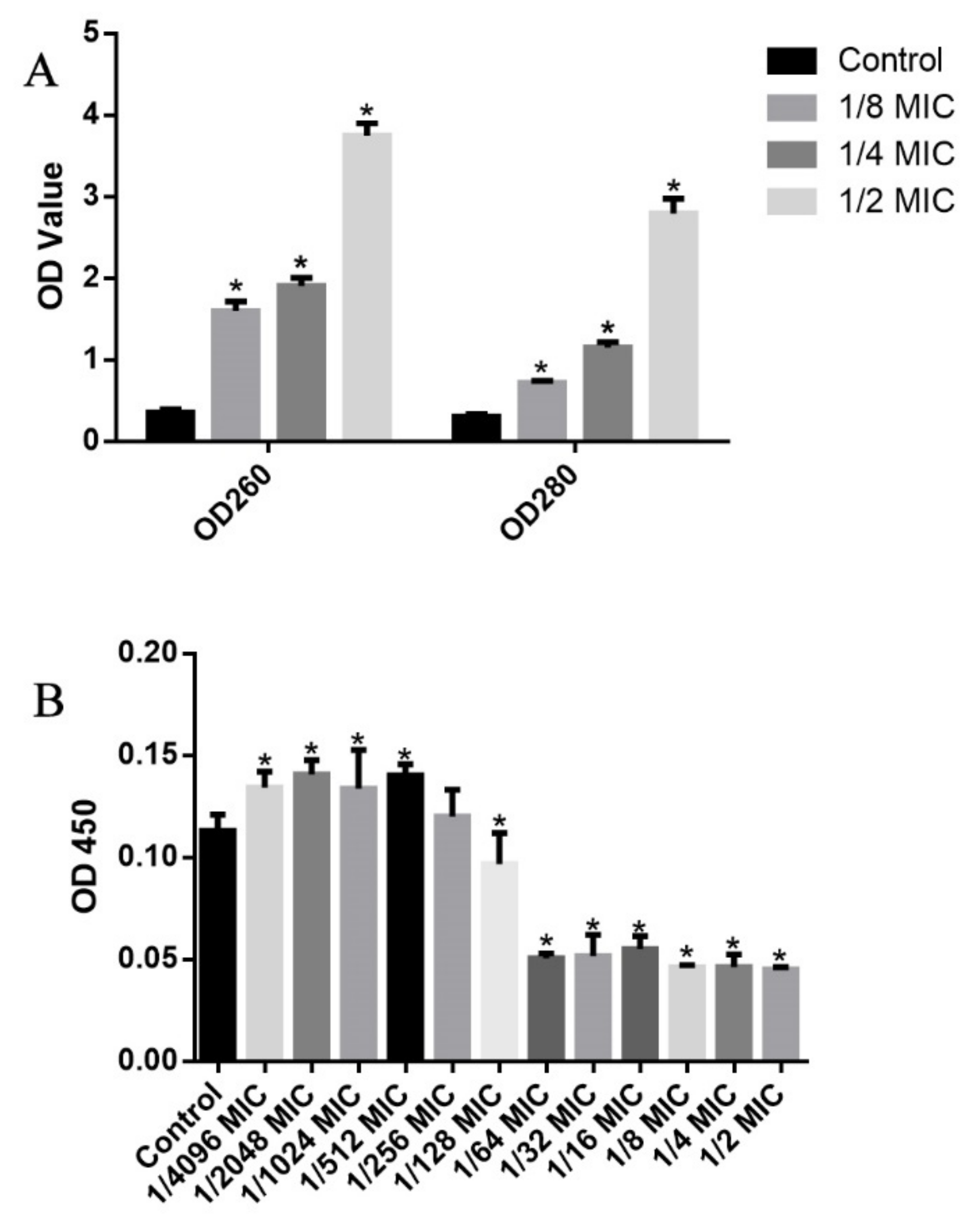
2.7. Leakages of Intracellular DNA and Protein
2.8. L. cubeba Essential Oil (LCEO) on Biofilm Formation
2.9. Fractional Inhibitory Concentration Index (FICI)
2.10. Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
2.11. Statistical Analysis of Quantitative Real-Time PCR and Opioid Addiction (OD) Value
3. Results and Discussion
3.1. MIC Values
3.2. Growth Curve and Time Kill Curve
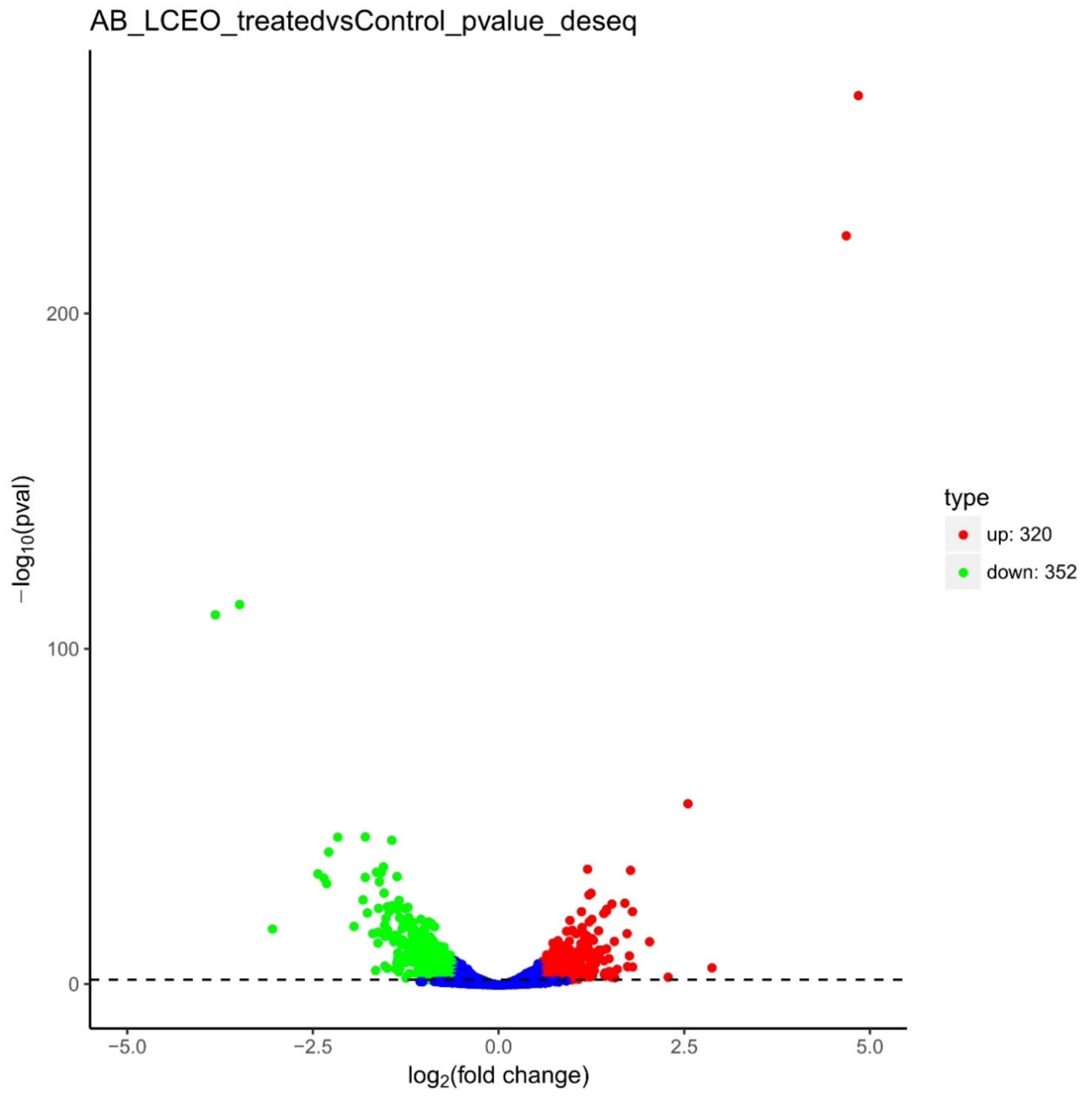
3.3. General Transcriptome Information
3.4. Differentially Expressed Genes in LCEO-Treated A. Baumannii 17
3.5. Gene Ontology (GO) Functional Enrichment Analysis
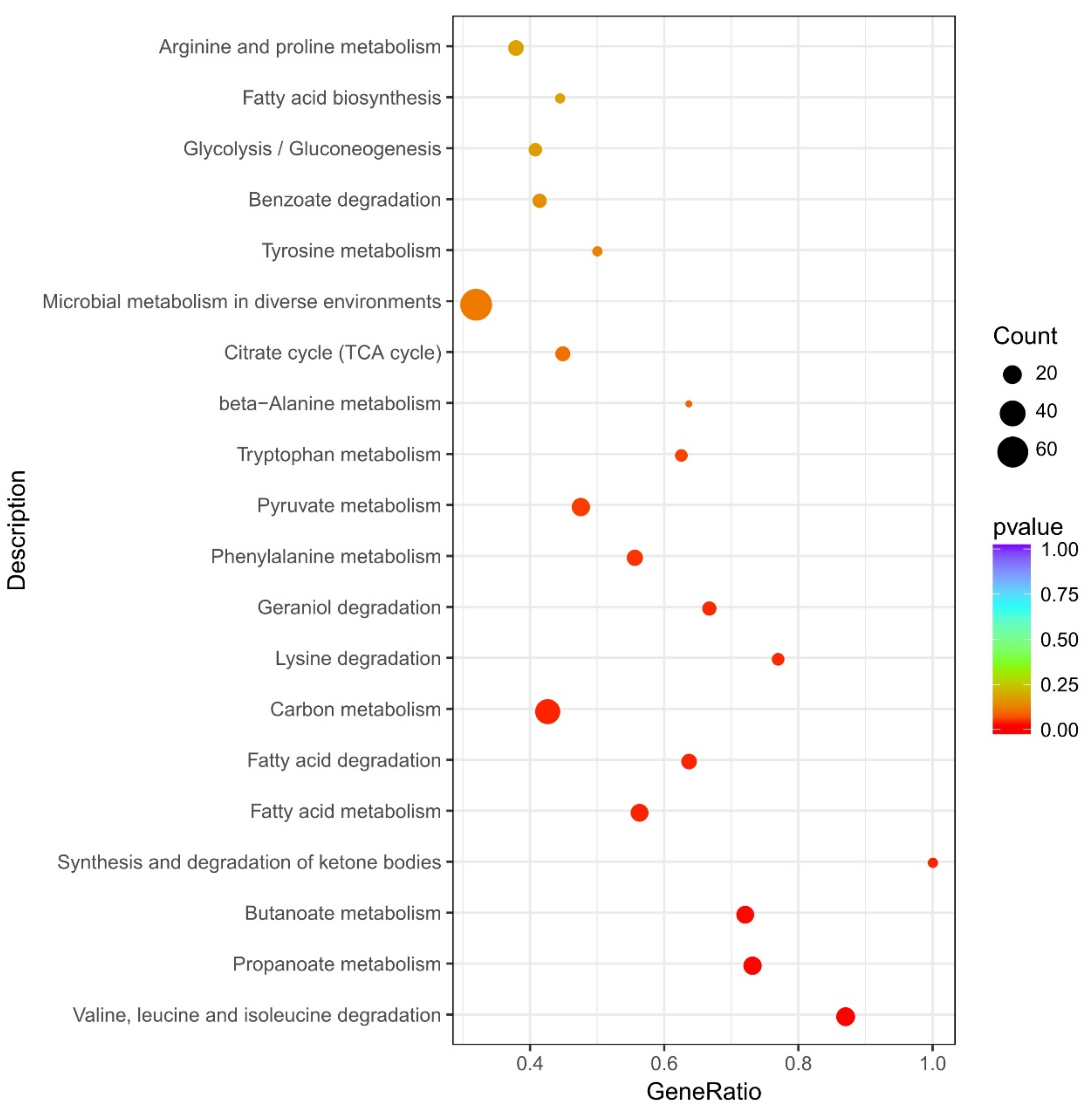
3.6. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
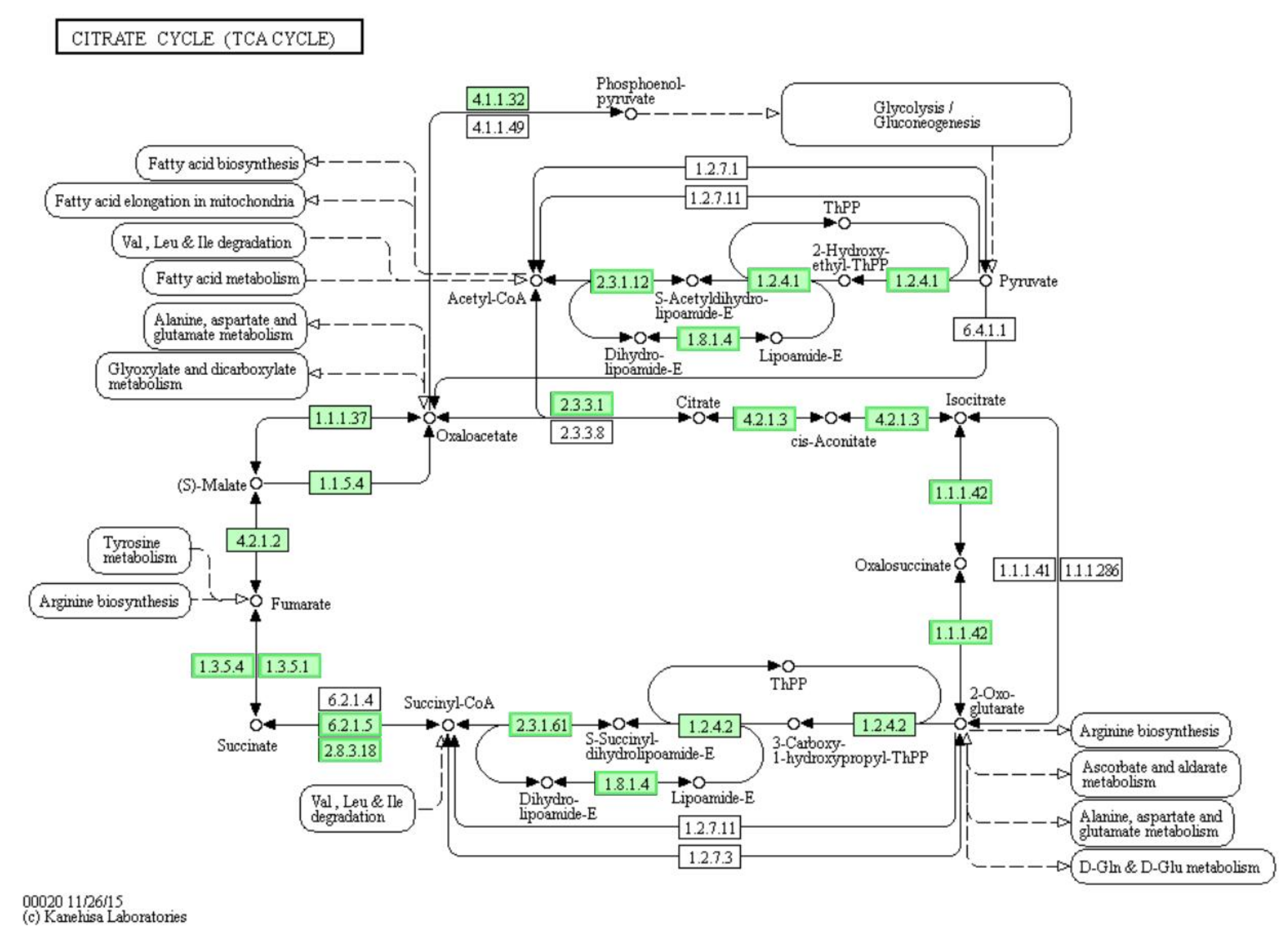
3.7. Differentially Expressed Genes (DEGs) Involved in “Citrate Cycle (TCA Cycle)” Pathway
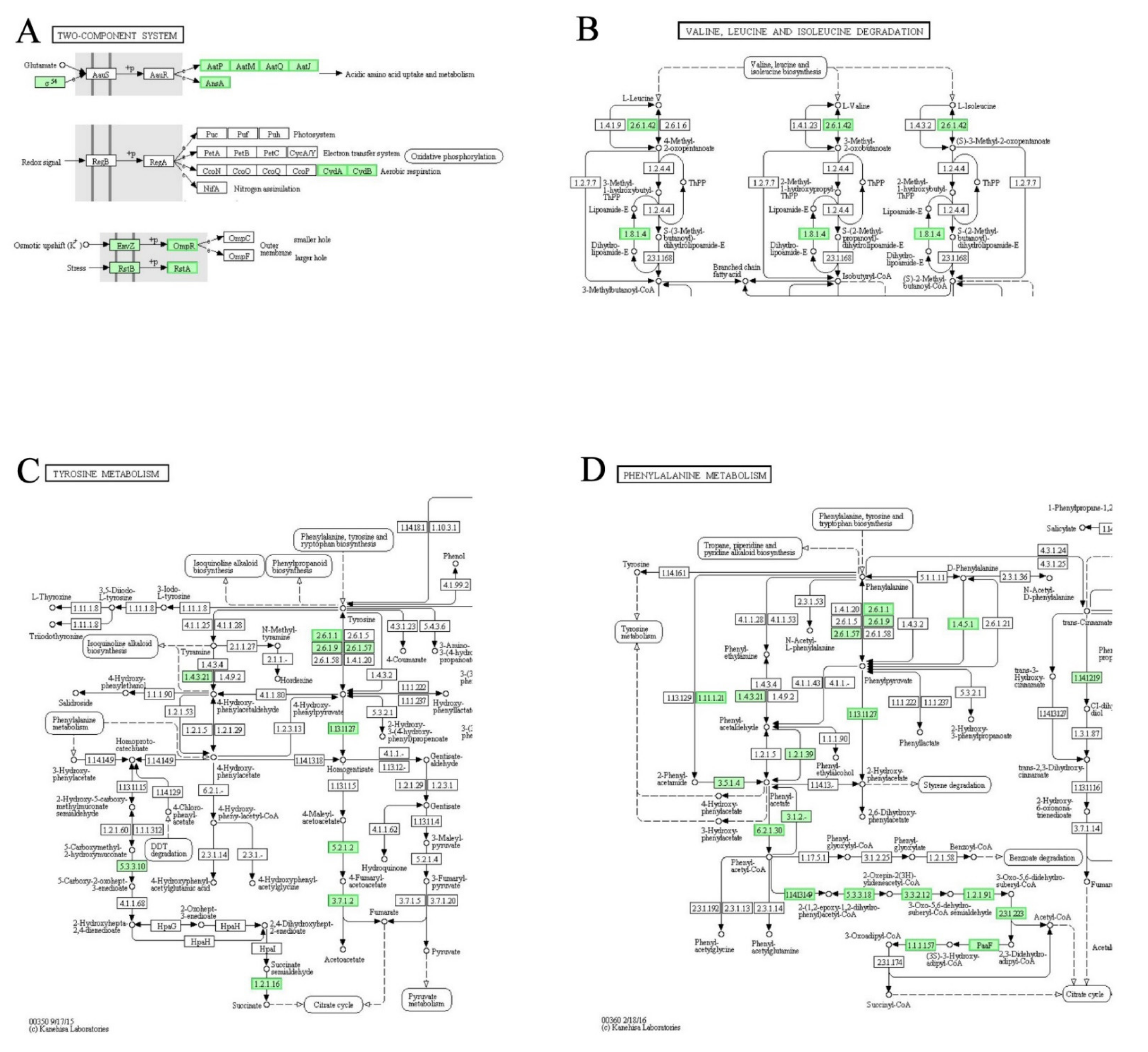
3.8. DEGs Involved in “Two-Component System”, “Valine, Leucine and Isoleucine Degradation”, “Tyrosine Metabolism” and “Phenylalanine Metabolism” Pathways
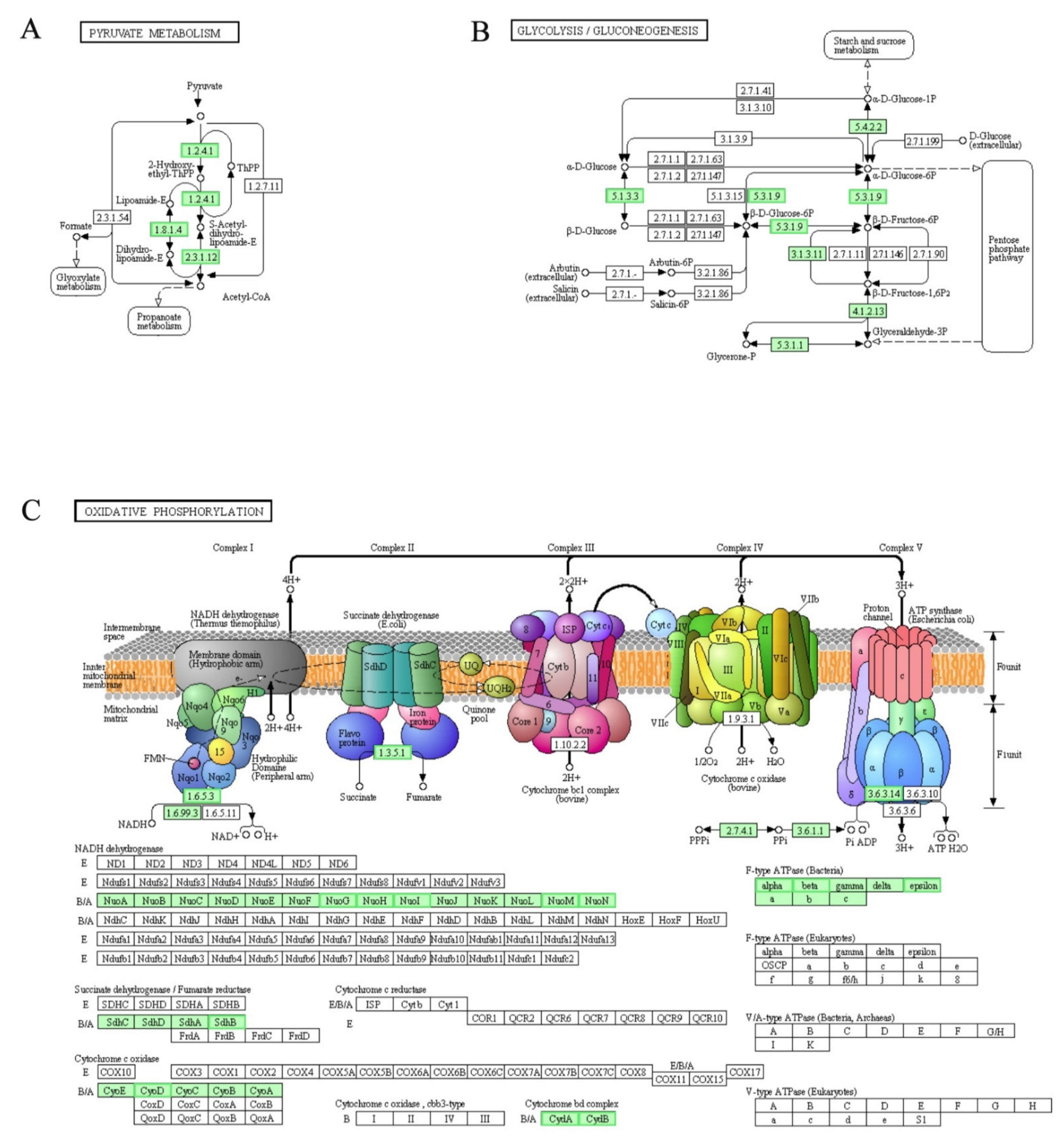
3.9. DEGs Involved in “Two-Component System”, “Pyruvate Metabolism”, “Glycolysis/Gluconeogenesis” and “Oxidative Phosphorylation” Pathways
3.10. DEGs Involved in “Synthesis and Degradation of Ketone Bodies” Pathway
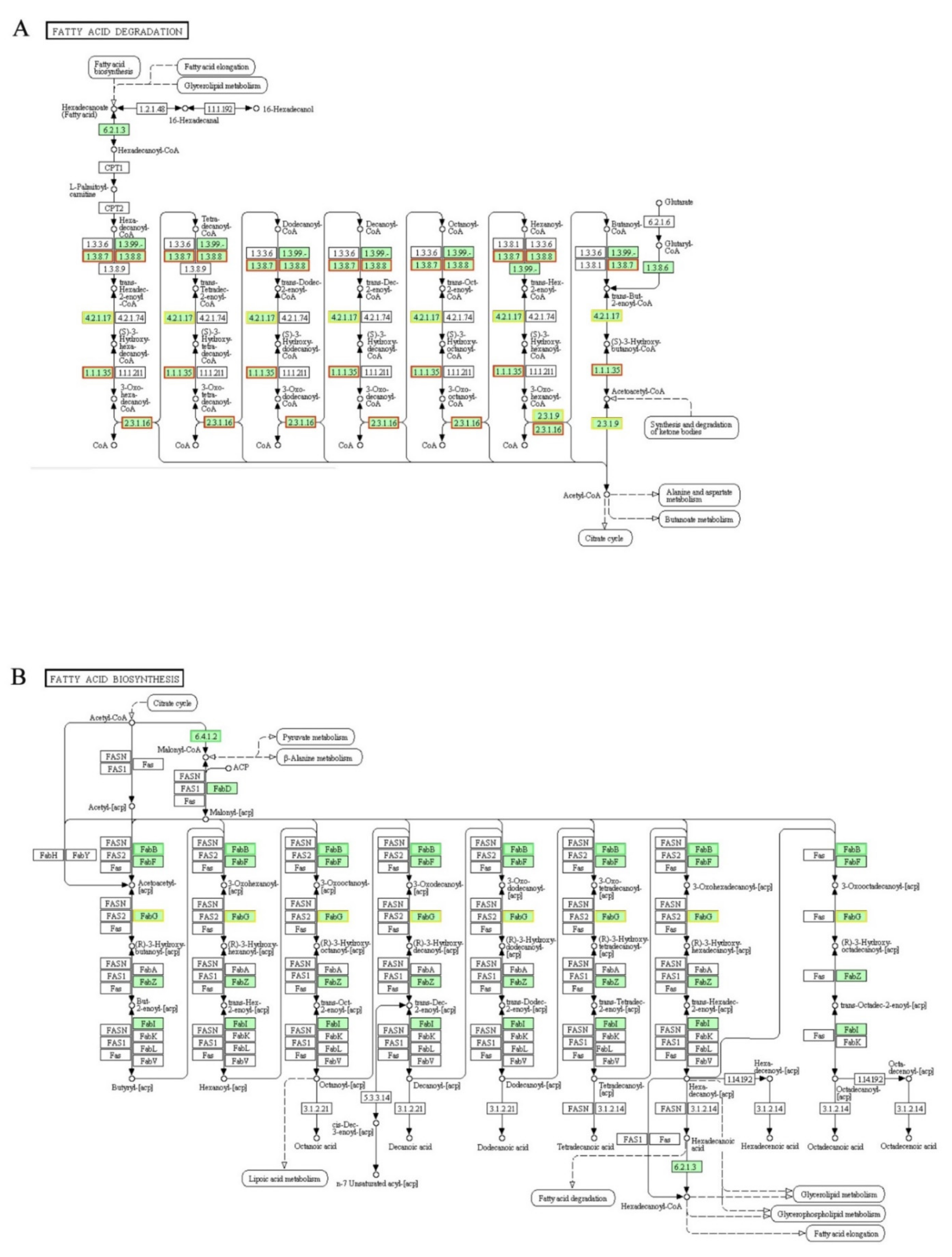
3.11. DEGs Involved in “Fatty Acid Degradation” and “Fatty Acid Biosynthesis” Pathways
3.12. Cell Membrane Damage and DEGs Involved in “Two-Component System”, “Histidine Metabolism”, “Peptidoglycan Biosynthesis” Pathways
3.13. Effects of LCEO on Biofilm Formation and FICI Values
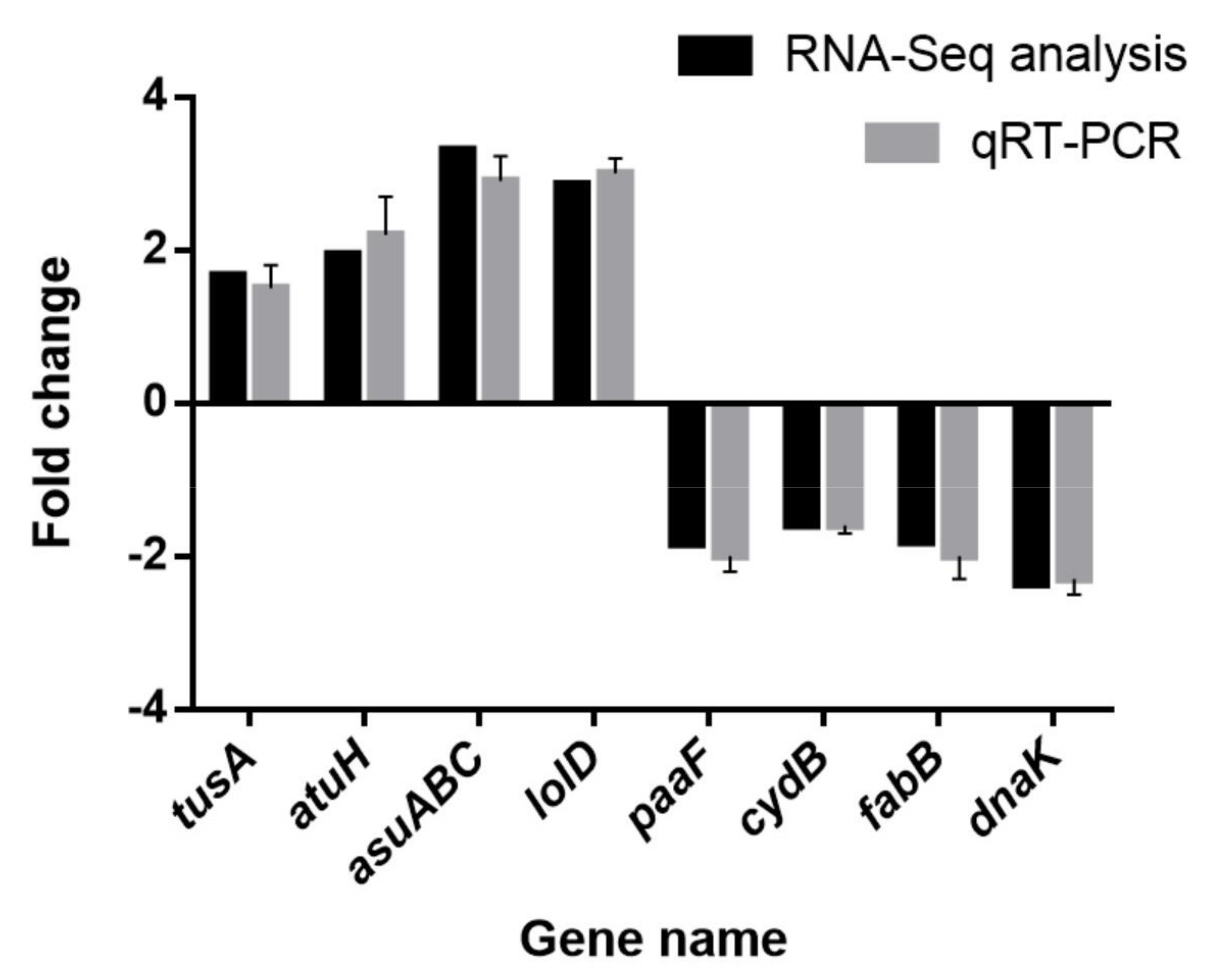
3.14. RT-PCR Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Meaning |
| LC | Litsea cubeba L. |
| LCEO | Litsea cubeba L. essential oil |
| MIC | Minimum Inhibitory Concentration |
| FICI | Fractional inhibitory concentration index |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| DEGs | Differentially expressed genes |
| A. baumannii | Acinetobacter baumannii |
| TCA cycle | Tricarboxylic acid cycle |
| TSA | Tryptone soy agar |
| TSB | Tryptic soy broth |
| CFU | Colony forming units |
| OD | Opioid addiction |
References
- Liu, T.-T.; Yang, T.-S. Antimicrobial impact of the components of essential oil of Litsea cubeba from Taiwan and antimicrobial activity of the oil in food systems. Int. J. Food Microbiol. 2012, 156, 68–75. [Google Scholar] [CrossRef]
- Ho, C.-L.; Jie-Ping, O.; Liu, Y.-C.; Hung, C.-P.; Tsai, M.-C.; Liao, P.-C.; Wang, E.I.-C.; Chen, Y.-L.; Su, Y.-C. Compositions and in vitro Anticancer activities of the Leaf and Fruit Oils of Litsea cubeba from Taiwan. Nat. Prod. Commun. 2010, 5, 617–620. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, Y.; Han, X.; Si, L.; Wu, Q.; Lin, L. Biology and chemistry ofLitsea cubeba, a promising industrial tree in China. J. Essent. Oil Res. 2013, 25, 103–111. [Google Scholar] [CrossRef]
- Van Nguyen, H.; Caruso, D.; Lebrun, M.; Nguyen, N.T.; Trinh, T.T.; Meile, J.; Chu, S.K.; Sarter, S. Antibacterial activity of Litsea cubeba (Lauraceae, May Chang) and its effects on the biological response of common carp Cyprinus carpio challenged with Aeromonas hydrophila. J. Appl. Microbiol. 2016, 121, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jiang, Z.-T.; Li, R. Complexation and molecular microcapsules of Litsea cubeba essential oil with β-cyclodextrin and its derivatives. Eur. Food Res. Technol. 2009, 228, 865–873. [Google Scholar] [CrossRef]
- Zhili, J.; Yasmin, A.; Rod, B.; Xing, Z.; Isman, M.B. Comparative toxicity of essential oils of Litsea pungens and Litsea cubeba and blends of their major constituents against the cabbage looper, Trichoplusia ni. J. Agric. Food Chem. 2009, 57, 4833–4837. [Google Scholar] [CrossRef]
- She, Q.-H.; Li, W.-S.; Jiang, Y.-Y.; Wu, Y.-C.; Zhou, Y.-H.; Zhang, L. Chemical composition, antimicrobial activity and antioxidant activity of Litsea cubeba essential oils in different months. Nat. Prod. Res. 2020, 34, 3285–3288. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-K.; Choi, E.-M.; Lee, J.H. Antioxidant activity of Litsea cubeba. Fitoterapia 2005, 76, 684–686. [Google Scholar] [CrossRef]
- Seo, S.-M.; Kim, J.; Lee, S.-G.; Shin, C.-H.; Shin, S.-C.; Park, I.-K. Fumigant Antitermitic Activity of Plant Essential Oils and Components from Ajowan (Trachyspermum ammi), Allspice (Pimenta dioica), Caraway (Carum carvi), Dill (Anethum graveolens), Geranium (Pelargonium graveolens), and Litsea (Litsea cubeba) Oils against Japanese Termite (Reticulitermes speratus Kolbe). J. Agric. Food Chem. 2009, 57, 6596–6602. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y. Chemical Composition and Antibacterial Activity of Essential Oils from Different Parts ofLitsea cubeba. Chem. Biodivers. 2010, 7, 229–235. [Google Scholar] [CrossRef]
- Hu, W.; Li, C.; Dai, J.; Cui, H.; Lin, L. Antibacterial activity and mechanism of Litsea cubeba essential oil against methicillin-resistant Staphylococcus aureus (MRSA). Ind. Crops Prod. 2019, 130, 34–41. [Google Scholar] [CrossRef]
- Li, W.-R.; Shi, Q.-S.; Liang, Q.; Xie, X.-B.; Huang, X.-M.; Chen, Y.-B. Antibacterial Activity and Kinetics of Litsea cubeba Oil on Escherichia coli. PLoS ONE 2014, 9, e110983. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Lebrun, M.; Caruso, D.; Chu-Ky, S.; Sarter, S.; Van Nguyen, H.; Meile, J.-C. Litsea cubeba leaf essential oil from Vietnam: Chemical diversity and its impacts on antibacterial activity. Lett. Appl. Microbiol. 2018, 66, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Dalsgaard, A.; Olsen, J.E. Phenotypic characterization and antibiotic resistance of Acinetobacter spp. isolated from aquatic sources. J. Appl. Microbiol. 1999, 87, 659–667. [Google Scholar] [CrossRef]
- Antunes, L.C.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurawski, D.V.; Banerjee, J.; Alamneh, Y.A.; Shearer, J.P.; Demons, S.T. Skin and Soft Tissue Models for Acinetobacter baumannii Infection. Methods Mol. Biol. 2019, 271–287. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Adukwu, E.C.; Bowles, M.; Edwards-Jones, V.; Bone, H. Antimicrobial activity, cytotoxicity and chemical analysis of lemongrass essential oil (Cymbopogon flexuosus) and pure citral. Appl. Microbiol. Biotechnol. 2016, 100, 9619–9627. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Chen, Y.; Zhang, G.; Sun, J.; Guo, L.; Jiang, M.; Ou, B.; Zhang, W.; Si, H. Transcriptomic Analysis of Staphylococcus aureus Under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, L.; Tariq, K.; Zhang, W.; Li, C.; Memon, F.Q.; Chai, B.; Li, Z.; Sun, J.; Chen, Y.; et al. The Antioxidant Polysaccharide from Semiaquilegia adoxoides (DC.) Makino Adjusts the Immune Response of Mice Infected by Bacteria. Evid. Based Complement. Altern. Med. 2020, 2020, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pan, J.; Ye, G.; Zhang, Q.; Wang, J.; Zhang, J.; Fang, J. In vitro studies of the antimicrobial effect of non-thermal plasma-activated water as a novel mouthwash. Eur. J. Oral Sci. 2017, 125, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.N.C.; Costa-Orlandi, C.B.; Bila, N.M.; Vaso, C.O.; Da Silva, R.A.M.; Mendes-Giannini, M.J.S.; Taylor, M.L.; Fusco-Almeida, A.M. Biofilm Formation by Histoplasma capsulatum in Different Culture Media and Oxygen Atmospheres. Front. Microbiol. 2020, 11, 1455. [Google Scholar] [CrossRef]
- Guo, N.; Ling, G.; Liang, X.; Jin, J.; Fan, J.; Qiu, J.; Song, Y.; Huang, N.; Wu, X.; Wang, X.; et al. In vitro synergy of pseudolaric acid B and fluconazole against clinical isolates of Candida albicans. Mycoses 2010, 54, e400–e406. [Google Scholar] [CrossRef]
- Li, Z.; Yin, H.; Chen, W.; Jiang, C.; Hu, J.; Xue, Y.; Yao, D.; Peng, Y.; Hu, X. Synergistic Effect of Pseudolaric Acid B with Fluconazole Against Resistant Isolates and Biofilm of Candida tropicalis. Infect. Drug Resist. 2020, 13, 2733–2743. [Google Scholar] [CrossRef]
- Moghimi, R.; Aliahmadi, A.; Rafati, H. Ultrasonic nanoemulsification of food grade trans-cinnamaldehyde: 1,8-Cineol and investigation of the mechanism of antibacterial activity. Ultrason. Sonochem. 2017, 35, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Terjung, N.; Löffler, M.; Gibis, M.; Hinrichs, J.; Weiss, J. Influence of droplet size on the efficacy of oil-in-water emulsions loaded with phenolic antimicrobials. Food Funct. 2012, 3, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Xu, B.; Zhang, G.; Lv, F.; Wang, Y.; Ma, M.; Si, H. Antibacterial Activity and Mechanism of Litsea cubeba L. Essential Oil Against Acinetobacter baumannii. Nat. Prod. Commun. 2021, 16. [Google Scholar] [CrossRef]
- Zheng, K.; Li, W.; Fu, B.; Fu, M.; Ren, Q.; Yang, F.; Qin, C. Physical, antibacterial and antioxidant properties of chitosan films containing hardleaf oatchestnut starch and Litsea cubeba oil. Int. J. Biol. Macromol. 2018, 118, 707–715. [Google Scholar] [CrossRef]
- Nakagami, K. [Studies on the shunt pathway of TCA cycle in avian type of tubercle bacillus. I. Citrate degradation]. Kekkaku [Tuberculosis] 1957, 32, 547–550. [Google Scholar]
- Atkinson, D.E. Citrate and the citrate cycle in the regulation of energy metabolism. Biochem. Soc. Symp. 1968, 27, 23–40. [Google Scholar]
- Goel, A.; Domach, M.M.; Hanley, W.; Lee, J.W.; Ataai, M.M. Coordination of Glycolysis and TCA Cycle Reaction Networks. Ann. N. Y. Acad. Sci. 1996, 782, 1–16. [Google Scholar] [CrossRef]
- Fukuchi, K.; Kasahara, Y.; Asai, K.; Kobayashi, K.; Moriya, S.; Ogasawara, N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 2000, 146, 1573–1583. [Google Scholar] [CrossRef] [Green Version]
- Pallegar, P.; Canuti, M.; Langille, E.; Peña-Castillo, L.; Lang, A.S. A Two-Component System Acquired by Horizontal Gene Transfer Modulates Gene Transfer and Motility via Cyclic Dimeric GMP. J. Mol. Biol. 2020, 432, 4840–4855. [Google Scholar] [CrossRef]
- Loessner, M.J.; Gaeng, S.; Wendlinger, G.; Maier, S.K.; Scherer, S. The two-component lysis system ofStaphylococcus aureusbacteriophage Twort: A large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 1998, 162, 265–274. [Google Scholar] [CrossRef]
- Stibftz, S.; Aaronson, W.; Monackt, D.; Falkowt, S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nat. Cell Biol. 1989, 338, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Meador, W.E.; Quiocho, F.A. Preliminary crystallographic data for a leucine, isoleucine, valine-binding protein from Escherichia coli K12. J. Mol. Biol. 1978, 123, 485–498. [Google Scholar] [CrossRef]
- Block, K.; Harper, A. Valine metabolism in vivo: Effects of high dietary levels of leucine and isoleucine. Metabolism 1984, 33, 559–566. [Google Scholar] [CrossRef]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trchounian, A.; Ohanjanyan, Y.; Bagramyan, K.; Vardanian, V.; Zakharyan, E.; Vassilian, A.; Davtian, M. Relationship of the Escherichia coli TrkA system of potassium ion uptake with the F0F1-ATPase under growth conditions without anaerobic or aerobic respiration. Biosci. Rep. 1998, 18, 143–154. [Google Scholar] [CrossRef]
- Tran, Q.H.; Unden, G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. JBIC J. Biol. Inorg. Chem. 1998, 251, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hulett, F.M. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: CtaA promoter regulation. Mol. Microbiol. 2000, 37, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Guest, J.R.; Angier, S.J.; Russell, G.C. Structure, Expression, and Protein Engineering of the Pyruvate Dehydrogenase Complex of Escherichia coli. Ann. N. Y. Acad. Sci. 1989, 573, 76–99. [Google Scholar] [CrossRef] [PubMed]
- Sugden, M.C.; Holness, M.J. The Role of Regulation of Tissue Pyruvate Dehydrogenase Complex Activity during the Starved-to-Fed Transition. Ann. N. Y. Acad. Sci. 1989, 573, 314–336. [Google Scholar] [CrossRef]
- Gol’dshtein, B.N.; Kornilov, V.V. Symmetry-regulated dynamics of multi-enzyme complexes. A model of a pyruvate dehydrogenase complex from Escherichia coli. Mol. Biol. 1988, 22, 538–548. [Google Scholar]
- Dallo, S.F.; Kannan, T.R.; Blaylock, M.W.; Baseman, J.B. Elongation factor Tu and E1 β subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 2002, 46, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, J.; Kawamori, R. Gluconeogenesis and glycolysis. Nihon Rinsho Jpn. J. Clin. Med. 2002, 60, 121–128. [Google Scholar]
- Gibson, F. The introduction of Escherichia coli and biochemical genetics to the study of oxidative phosphorylation. Trends Biochem. Sci. 2000, 25, 342–344. [Google Scholar] [CrossRef]
- Fukao, T.; Mitchell, G.; Sass, J.O.; Hori, T.; Orii, K.; Aoyama, Y. Ketone body metabolism and its defects. J. Inherit. Metab. Dis. 2014, 37, 541–551. [Google Scholar] [CrossRef]
- Lundgren, B.R.; Harris, J.R.; Scheel, R.A.; Sarwar, Z.; Nomura, C.T. The metabolism of (R)-3-hydroxybutyrate is regulated by the enhancer-binding protein PA2005 and the alternative sigma factor RpoN in Pseudomonas aeruginosa PAO1. Microbiology 2015, 161, 2232–2242. [Google Scholar] [CrossRef]
- Potezny, N.; Atkinson, E.R.; Rofe, A.M.; Conyers, R.A. The inhibition of bacterial cell growth by ketone bodies. Aust. J. Exp. Biol. Med. Sci. 1981, 59, 639–649. [Google Scholar] [CrossRef]
- Parekh, V.R.; Traxler, R.W.; Sobek, J.M. N-Alkane oxidation enzymes of a pseudomonad. Appl. Environ. Microbiol. 1977, 33, 881–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatesan, R.; Sah-Teli, S.K.; Awoniyi, L.O.; Jiang, G.; Prus, P.; Kastaniotis, A.J.; Hiltunen, J.K.; Wierenga, R.K.; Chen, Z. Insights into mitochondrial fatty acid synthesis from the structure of heterotetrameric 3-ketoacyl-ACP reductase/3R-hydroxyacyl-CoA dehydrogenase. Nat. Commun. 2014, 5, 4805. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, R.D.S.; Balbino, B.L.; Andrade, G.; Dias, M.C.P.S.; Alvarenga, T.A.; Pedroso, R.C.N.; Pimenta, L.P.; Lucarini, R.; Pauletti, P.M.; Januário, A.H.; et al. In Vitro and In Vivo Anti-Candida spp. Activity of Plant-Derived Products. Plants 2019, 8, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzyśko-Łupicka, T.; Sokół, S.; Piekarska-Stachowiak, A.A. Evaluation of Fungistatic Activity of Eight Selected Essential Oils on Four Heterogeneous Fusarium Isolates Obtained from Cereal Grains in Southern Poland. Molecules 2020, 25, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Shi, W.; Miklossy, J.; Tauxe, G.M.; McMeniman, C.J.; Zhang, Y. Identification of Essential Oils with Strong Activity against Stationary Phase Borrelia burgdorferi. Antibiotics 2018, 7, 89. [Google Scholar] [CrossRef] [Green Version]
- Van Heijenoort, J. Recent advances in the formation of the bacterial peptidoglycan monomer unit (1985 to 2000). Nat. Prod. Rep. 2001, 18, 503–519. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Hou, Y.; Soteyome, T.; Zeng, B.; Su, J.; Li, L.; Li, B.; Chen, D.; Li, Y.; et al. Transcriptomics Study on Staphylococcus aureus Biofilm Under Low Concentration of Ampicillin. Front. Microbiol. 2018, 9, 2413. [Google Scholar] [CrossRef] [Green Version]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; O Sintim, H. Biofilm formation mechanisms and targets for developing antibiofilm agents. Futur. Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]



| Composition | Ratio (%) |
|---|---|
| α-Citral | 38.28 |
| β-Citral | 29.29 |
| Cinene | 16.5348 |
| Eudesmol | 2.56 |
| Citronellal | 2.2383 |
| Trans-Verbenol | 2.06 |
| Aromatic alcohol | 1.86 |
| β-Pinene | 1.5104 |
| Cis-Verbenol | 1.3771 |
| α-Vinyl acetate | 1.0945 |
| Geraniol | 0.93 |
| α-Pinene | 0.7307 |
| 2-Methyl-2-Hexen-6-One | 0.6994 |
| Gene Name | Primer Sequence (5′->3′) | NCBI-Protein ID | Products (Base Pairs) |
|---|---|---|---|
| tusA | AGATGTTGTTGAAGTGTT CCAGATAACGGTATTCTT | ABO10836 | 137 |
| atuH | AATGATTGGTAATGGTATG AACTGGATAATGGCATAT | ABO10859 | 174 |
| asuABC | GGAGTGATGATGCTAGGT TAGAATAGGAAGGATGCCATA | ABO10525 | 96 |
| lolD | AGCAGACAGTCAATATAAGT GGTCCATACGATGAGATAG | ABO13028 | 80 |
| paaF | AAGAAGAATTACTTGAACA TATTGAATGGAACATACG | ABO10593 | 150 |
| cydB | AATATCATTCCACCATCA ATAACCTTCATCACCTAC | ABO11861 | 165 |
| fabB | GGAACGAGTTGTCATCAC GTTATAGCGAATGCCAGAG | ABO10573 | 106 |
| dnaK | TCTACTGCTGCTGATAAC GATGTCACCTAACTGGAA | ABO13363 | 108 |
| Component | A. baumannii 4 | A. baumannii 20 | A. baumannii 17 |
|---|---|---|---|
| Ceftriaxone Sodium | 3.2 | 1.6 | 0.8 |
| Amoxicillin | 0.8 | 0.8 | 1.6 |
| Berberine | 0.0015625 | 0.003125 | 0.003125 |
| Neomycin | 0.4 | 0.2 | 3.2 |
| Levofloxacin | 0.0125 | 0.025 | 0.025 |
| Fosfomycin | 3.2 | 3.2 | 0.4 |
| Colistin | 0.0015625 | 0.0015625 | 0.0015625 |
| Mequindox | 0.1 | 0.2 | 0.1 |
| Clinfloxacin | 0.025 | 0.025 | 0.025 |
| Sulfamonomethoxine | 6.4 | 6.4 | 3.2 |
| Gatifloxacin | 0.003125 | 0.025 | 0.025 |
| Amikacin | 0.8 | 6.4 | 3.2 |
| Ceftazidime | 0.2 | 0.025 | 0.1 |
| Lincomycin | 0.4 | 0.4 | 3.2 |
| Ceftiofur Sodium | 3.2 | 0.8 | 0.4 |
| Florfenicol | 0.32 | 0.16 | 0.32 |
| Azithromycin | 0.8 | 0.4 | 3.2 |
| Cefotaxime Sodium | 0.0125 | 0.2 | 0.1 |
| Rifampin | 0.0125 | 0.05 | 0.025 |
| Meropenem | 0.2 | 0.1 | 0.2 |
| Ceftriaxone Sodium | 3.2 | 1.6 | 0.8 |
| LCEO | 1.08 | 1.08 | 1.08 |
| Tween 80 | >108 | >108 | >108 |
| Drugs | MIC Alone | MIC Combined | FICI | Interpretation |
|---|---|---|---|---|
| LCEO | 1.08 mg/mL | 0.27 mg/mL | 0.75 | ADD |
| Tetracycline | 1.28 mg/mL | 0.64 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.27 mg/mL | 1.25 | IND |
| Meropenem | 0.32 mg/mL | 0.32 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.27 mg/mL | 0.375 | SYN |
| Chloramphenicol | 0.32 mg/mL | 0.04 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.27 mg/mL | 1.25 | IND |
| Amikacin | 1.28 mg/mL | 1.28 mg/mL | ||
| LCEO | 1.08 mg/mL | 1.08 mg/mL | 2 | IND |
| Ampicillin | 0.32 mg/mL | 0.32 mg/mL | ||
| LCEO | 1.08 mg/mL | 1.08 mg/mL | 2 | IND |
| Kanamycin | 0.32 mg/mL | 0.32 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.27 mg/mL | 1.25 | IND |
| Ciprofloxacin | 0.64 mg/mL | 0.64 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.54 mg/mL | 1.5 | IND |
| Fosfomycin | 0.64 mg/mL | 0.64 mg/mL | ||
| LCEO | 1.08 mg/mL | 1.08 mg/mL | 2 | IND |
| Streptomycin sulfate | 1.28 mg/mL | 1.28 mg/mL | ||
| LCEO | 1.08 mg/mL | 0.54 mg/mL | 2.5 | IND |
| Ceftiofur | 0.64 mg/mL | 1.28 mg/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Hao, K.; Jiang, M.; Memon, F.U.; Guo, L.; Zhang, G.; Liu, T.; Wu, X.; Si, H. Transcriptomic Analysis of Drug-Resistance Acinetobacter baumannii under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing. Genes 2021, 12, 1003. https://doi.org/10.3390/genes12071003
Yang Y, Hao K, Jiang M, Memon FU, Guo L, Zhang G, Liu T, Wu X, Si H. Transcriptomic Analysis of Drug-Resistance Acinetobacter baumannii under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing. Genes. 2021; 12(7):1003. https://doi.org/10.3390/genes12071003
Chicago/Turabian StyleYang, Yunqiao, Kaiyuan Hao, Mingsheng Jiang, Fareed Uddin Memon, Lei Guo, Geyin Zhang, Tian Liu, Xianshi Wu, and Hongbin Si. 2021. "Transcriptomic Analysis of Drug-Resistance Acinetobacter baumannii under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing" Genes 12, no. 7: 1003. https://doi.org/10.3390/genes12071003
APA StyleYang, Y., Hao, K., Jiang, M., Memon, F. U., Guo, L., Zhang, G., Liu, T., Wu, X., & Si, H. (2021). Transcriptomic Analysis of Drug-Resistance Acinetobacter baumannii under the Stress Condition Caused by Litsea cubeba L. Essential Oil via RNA Sequencing. Genes, 12(7), 1003. https://doi.org/10.3390/genes12071003