In Silico Prediction of Transcription Factor Collaborations Underlying Phenotypic Sexual Dimorphism in Zebrafish (Danio rerio)
Abstract
1. Introduction
2. Materials and Methods
2.1. Candidate Sex and Colour Genes
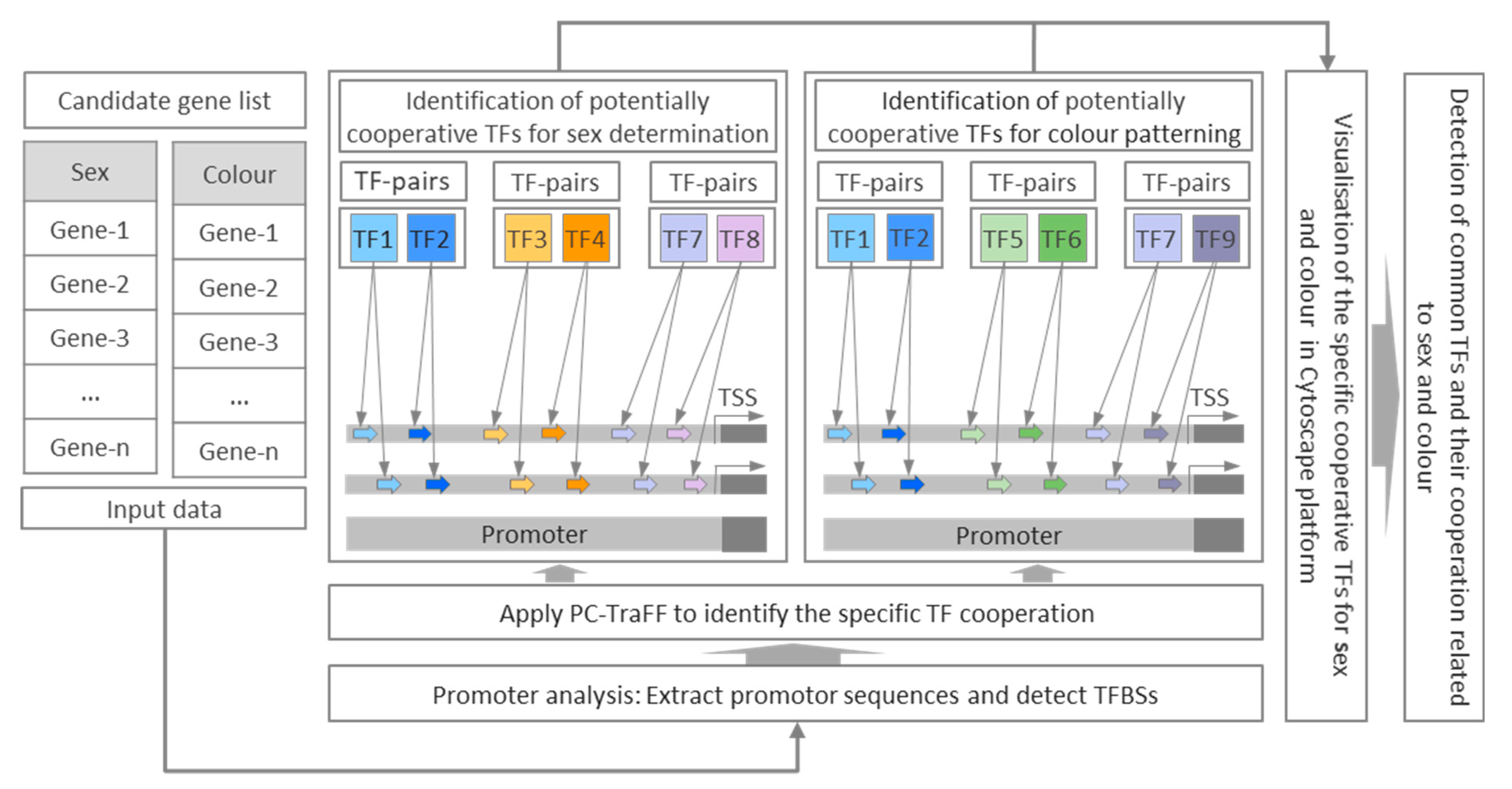
2.2. Identification of Potentially Cooperative Transcription Factors
3. Results and Discussion
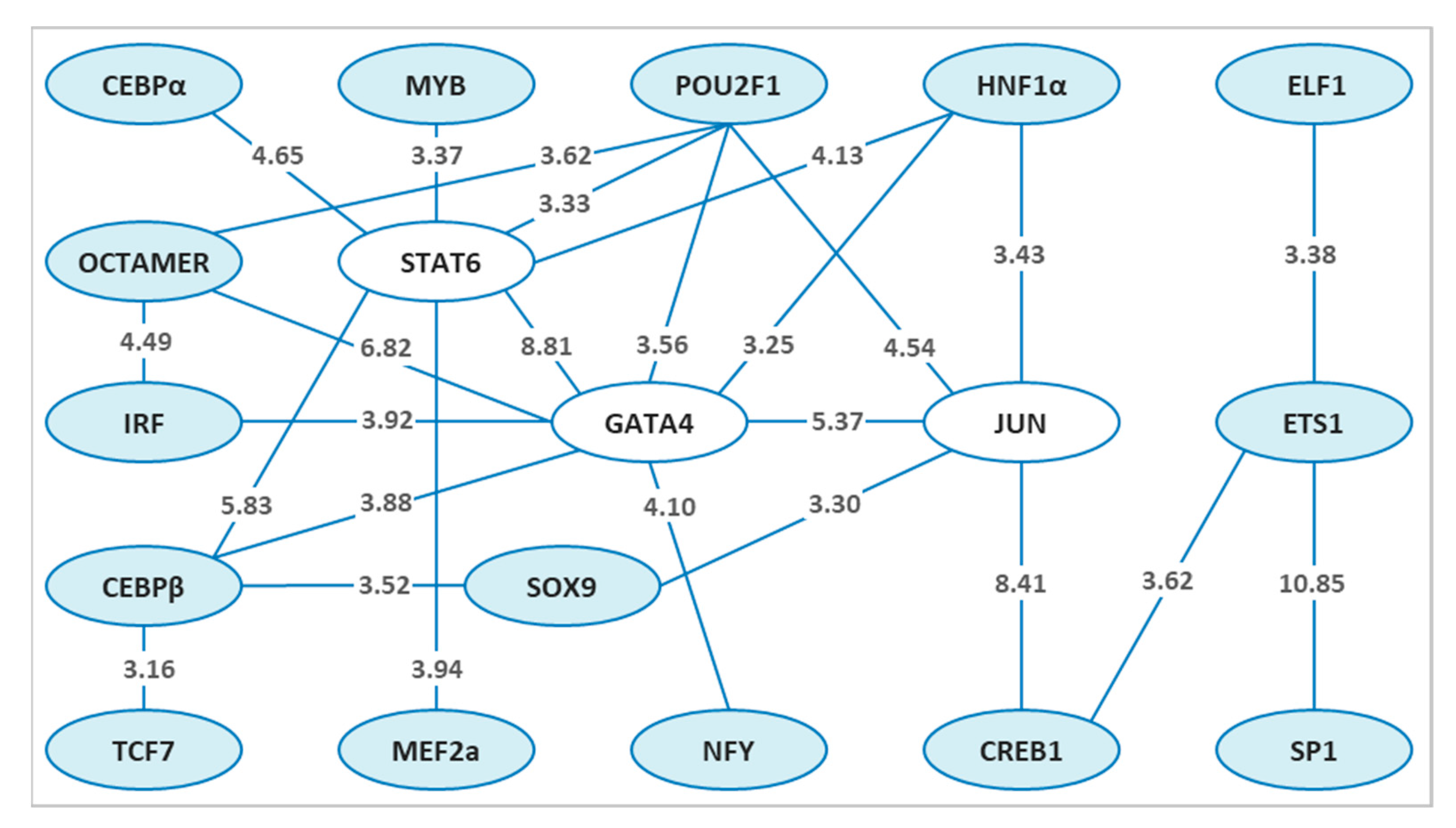
3.1. Potentially Cooperative TF Pairs Directing Sex Determination in Zebrafish
3.2. Potentially Cooperative TF Pairs Directing Colour Patterning in Zebrafish
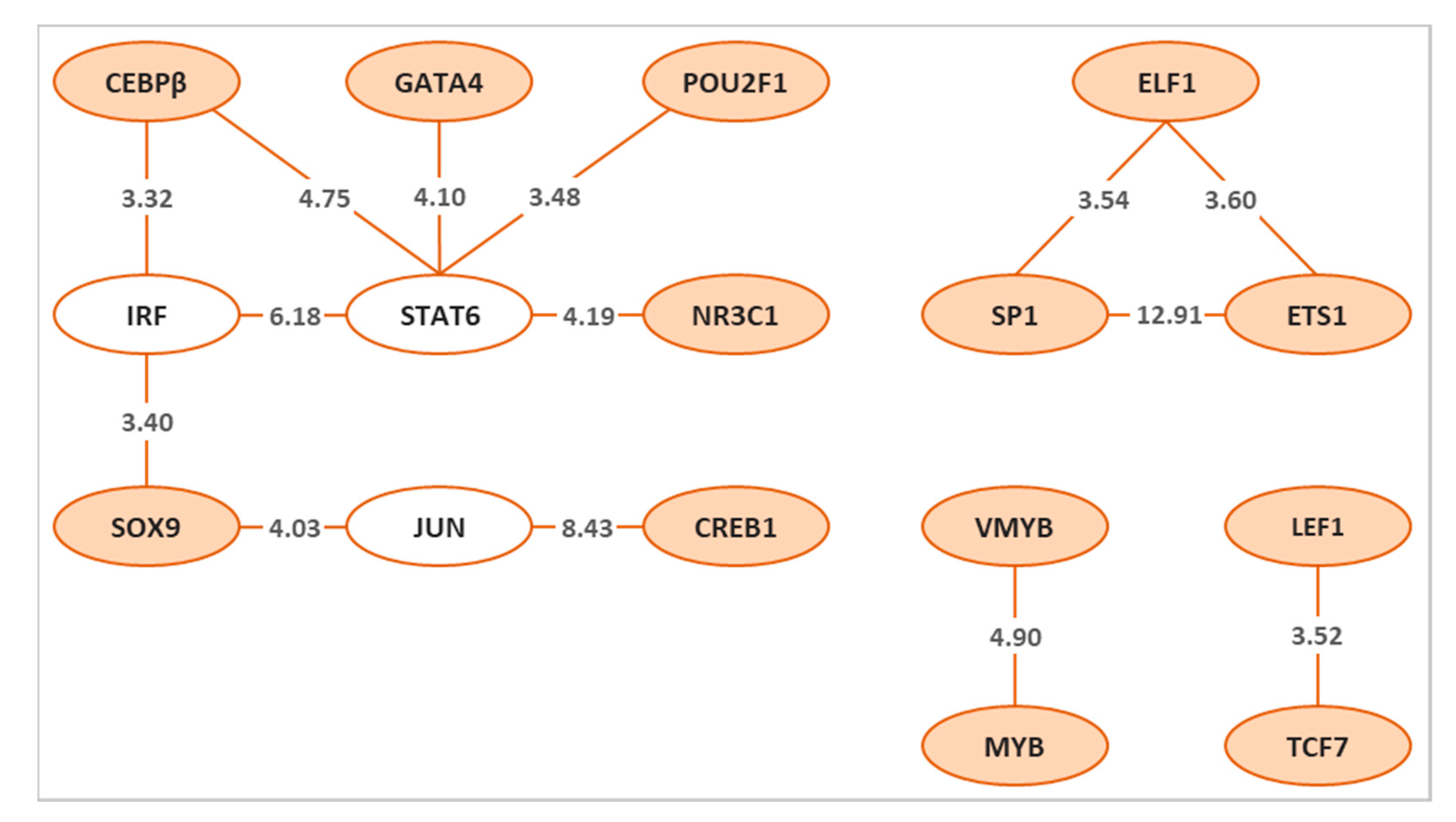
3.3. Identification of Common Cooperative TFs for Sex and Colour Genes in Zebrafish
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jim, H.L. Identification of Target Genes of an Erythroid Transcription Factor Complex Containing SCL (TAL1). Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2008; pp. 1–7. [Google Scholar]
- Inukai, S.; Kock, K.H.; Bulyk, M.L. Transcription factor-DNA binding: Beyond binding site motifs. Curr. Opin. Genet. Dev. 2017, 43, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Rinaldi, N.J.; Robert, F.; Odom, D.T.; Bar-Joseph, Z.; Gerber, G.K.; Hannett, N.M.; Harbison, C.T.; Thompson, C.M.; Simon, I.; et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 2002, 298, 799–804. [Google Scholar] [CrossRef]
- Thakurta, D.G. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006, 34, 3585–3598. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Robertson, D.L.; Peer, Y.V.D.; Oliver, S.G. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 2008, 33, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Parma, P.; Radi, O. Molecular mechanisms of sexual development. Sex. Dev. 2012, 6, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C.; Mank, J.E. Evolutionary perspectives on hermaphroditism in fishes. Sex. Dev. 2009, 3, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Dranow, D.B.; Hu, K.; Bird, A.M.; Lawry, S.T.; Adams, M.T.; Sanchez, A.; Amatruda, J.F.; Draper, B.W. Bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 2016, 12, e1006323. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef]
- Vilain, E.; McCabe, E.R. Mammalian sex determination: From gonads to brain. Mol. Genet. Metab. 1998, 65, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Hughes, I.A. Minireview: Sex differentiation. Endocrinology 2001, 142, 3281–3287. [Google Scholar] [CrossRef]
- Arnold, A.P.; Burgoyne, P.S. Are XX and XY brain cells intrinsically different? Trends Endocrinol. Metab. 2004, 15, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Schadt, E.E.; Wang, S.; Wang, H.; Arnold, A.P.; Ingram-Drake, L.; Drake, T.A.; Lusis, A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- She, Z.Y.; Yang, W.X. Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 2017, 63, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, G.; Vilain, E. Mechanisms of disease: Transcription factors in sex determination—Relevance to human disorders of sex development. Nat. Clin. Pract. Endocrinol. Metab. 2006, 2, 231–238. [Google Scholar] [CrossRef]
- Wilson, C.A.; High, S.K.; McCluskey, B.M.; Amores, A.; Yan, Y.L.; Titus, T.A.; Anderson, J.L.; Batzel, P.; Carvan, M.J., III; Schartl, M.; et al. Wild sex in zebrafish: Loss of the natural sex determinant in domesticated strains. Genetics 2014, 198, 1291–1308. [Google Scholar] [CrossRef]
- Liew, W.C.; Bartfai, R.; Lim, Z.; Sreenivasan, R.; Siegfried, K.R.; Orban, L. Polygenic sex determination system in zebrafish. PLoS ONE 2012, 7, e34397. [Google Scholar] [CrossRef]
- Liew, W.C.; Orban, L. Zebrafish sex: A complicated affair. Brief. Funct. Genom. 2014, 13, 172–187. [Google Scholar] [CrossRef]
- Hosseini, S.; Ha, N.T.; Simianer, H.; Falker-Gieske, C.; Brenig, B.; Franke, A.; Hörstgen-Schwark, G.; Tetens, J.; Herzog, S.; Sharifi, A.R. Genetic mechanism underlying sexual plasticity and its association with colour patterning in zebrafish (Danio rerio). BMC Genom. 2019, 20, 341. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Brenig, B.; Tetens, J.; Sharifi, A.R. Phenotypic plasticity induced using high ambient temperature during embryogenesis in domesticated zebrafish. Reprod. Domest. Anim. 2019, 54, 435–444. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DMdomain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, M.; Shinomiya, A.; Kinoshita, M.; Suzuki, A.; Kobayashi, T.; Paul-Prasanth, B.; Lau, E.-L.; Hamaguchi, S.; Sakaizumi, M.; Nagahama, Y. DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. USA 2007, 104, 3865–3870. [Google Scholar] [CrossRef]
- Webster, K.A.; Schach, U.; Ordaz, A.; Steinfeld, J.S.; Draper, B.W.; Siegfried, K.R. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 2017, 422, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Zhang, Z.; Song, W.; Wong, Q.W.L.; Chen, W.; Shirgaonkar, N.; Ge, W. Roles of Figla/figla in Juvenile Ovary Development and Follicle Formation during Zebrafish Gonadogenesis. Endocrinology 2018, 159, 3699–3722. [Google Scholar] [CrossRef]
- King, A.C.; Gut, M.; Zenker, A.K. Shedding new light on early sex determination in zebrafish. Arch Toxicol. 2020, 94, 4143–4158. [Google Scholar] [CrossRef]
- Kossack, M.E.; Draper, B.W. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr. Top. Dev. Biol. 2019, 134, 119–149. [Google Scholar]
- Parichy, D.M.; Turner, J.M. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development 2003, 130, 817–833. [Google Scholar] [CrossRef]
- Lang, D.; Epstein, J.A. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum. Mol. Genet. 2003, 12, 937–945. [Google Scholar] [CrossRef]
- Minchin, J.E.N.; Hughes, S.M. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev. Biol. 2008, 317, 508–522. [Google Scholar] [CrossRef]
- Koludrovic, D.; Davidson, I. MITF, the Janus transcription factor of melanoma. Future Oncol. 2013, 9, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Wen, S.; Luo, C.; Zhang, Y.Q.; Tao, M.; Wang, D.W.; Deng, S.M.; Xiao, Y.M. Involvement of the mitfa gene in the development of pigment cell in Japanese ornamental (Koi) carp (Cyprinus carpio L.). Genet. Mol. Res. 2015, 14, 2775–2784. [Google Scholar] [CrossRef]
- Protas, M.E.; Patel, N.H. Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 2008, 24, 425–446. [Google Scholar] [CrossRef]
- Lin, S.J.; Foley, J.; Jiang, T.X.; Yeh, C.Y.; Wu, P.; Foley, A.; Yen, C.M.; Huang, Y.C.; Cheng, H.C.; Chen, C.F.; et al. Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science 2013, 340, 1442–1445. [Google Scholar] [CrossRef]
- Reudink, M.W.; McKellar, A.E.; Marini, K.L.; McArthur, S.L.; Marra, P.P.; Ratcliffe, L.M. Inter-annual variation in American redstart (Setophaga ruticilla) plumage colour is associated with rainfall and temperature during moult: An 11-year study. Oecologia 2015, 178, 161–173. [Google Scholar] [CrossRef]
- Darwin, C.R. The Descent of Man, and Selection in Relation to Sex; D. Appleton and Company: New York, NY, USA, 1889. [Google Scholar]
- Rashed, A.; Polak, M. Does male secondary sexual trait size reveal fertilization efficiency in Australian Drosophila bipectinata Duda (Diptera: Drosophilidae)? Biol. J. Linn. Soc. 2009, 98, 406–413. [Google Scholar] [CrossRef]
- Gronell, A.M. Visiting behaviour by females of the sexually dichromatic damselfish, Chrysiptera cyanea (Teleostei: Pomacentridae): A probable method of assessing male quality. Ethology 1989, 81, 89–122. [Google Scholar] [CrossRef]
- Kraak, S.B.M.; Bakker, T.C.M.; Mundwiler, B. Sexual selection in sticklebacks in the field: Correlates of reproductive, mating, and paternal success. Behav. Ecol. 1999, 10, 696–706. [Google Scholar] [CrossRef]
- Wacker, S.; Östlund-Nilsson, S.; Forsgren, E.; Newport, C.; Amundsen, T. Mate choice plasticity in a coral reef fish. Behav. Ecol. 2016, 27, 1331–1342. [Google Scholar] [CrossRef]
- Engeszer, R.E.; Ryan, M.J.; Parichy, D.M. Learned social preference in zebrafish. Curr. Biol. 2004, 14, 881–884. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, G.G.; Ryan, M.J. Assortative preferences for stripes in danios. Anim. Behav. 2005, 70, 1063–1066. [Google Scholar] [CrossRef]
- Ruhl, N.; McRobert, S.P. The effect of sex and shoal size on shoaling behaviour in Danio rerio. J. Fish Biol. 2005, 67, 1318–1326. [Google Scholar] [CrossRef]
- Snekser, J.L.; McRobert, S.P.; Murphy, C.E.; Clotfelter, E.D. Aggregation behaviour in wildtype and transgenic zebrafish. Ethology 2006, 112, 181–187. [Google Scholar] [CrossRef]
- Hutter, S.; Penn, D.J.; Magee, S.; Zala, S.M. Reproductive behaviour of wild zebrafish (Danio rerio) in large tanks. Behaviour 2010, 147, 641–660. [Google Scholar] [CrossRef]
- Singh, A.P.; Nüsslein-Volhard, C. Zebrafish stripes as a model for vertebrate colour pattern formation. Curr. Biol. 2015, 25, R81–R92. [Google Scholar] [CrossRef]
- Nüsslein-Volhard, C.; Singh, A.P. How fish colour their skin: A paradigm for development and evolution of adult patterns. Bioessays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Simianer, H.; Tetens, J.; Brenig, B.; Herzog, S.; Sharifi, A.R. Efficient phenotypic sex classification of zebrafish using machine learning methods. Ecol. Evol. 2019, 9, 13332–13343. [Google Scholar] [CrossRef] [PubMed]
- Hutter, S.; Zala, S.M.; Penn, D.J. Sex recognition in zebrafish (Danio rerio). J. Ethol. 2011, 29, 55–61. [Google Scholar] [CrossRef]
- Hutter, S.; Hettyey, A.; Penn, D.J.; Zala, S.M. Ephemeral Sexual Dichromatism in Zebrafish (Danio rerio). J. Ethol. 2012, 118, 1208–1218. [Google Scholar] [CrossRef]
- Seberg, H.E.; Otterloo, E.V.; Cornell, R.A. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017, 30, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Elworthy, S.; Lister, J.A.; Carney, T.J.; Raible, D.W.; Kelsh, R.N. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 2003, 130, 2809–2818. [Google Scholar] [CrossRef]
- Braasch, I.; Brunet, F.; Volff, J.N.; Schartl, M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol. Evol. 2009, 1, 479–493. [Google Scholar] [CrossRef]
- Sharma, E.; Künstner, A.; Fraser, B.A.; Zipprich, G.; Kottler, V.A.; Henz, S.R.; Weigel, D.; Dreyer, C. Transcriptome assemblies for studying sex-biased gene expression in the guppy, Poecilia reticulate. BMC Genom. 2014, 15, 400. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Z.; He, S. Both male-biased and female-biased genes evolve faster in fish genomes. Genome Biol. Evol. 2016, 8, 3433–3445. [Google Scholar] [CrossRef] [PubMed]
- Ranz, J.M.; Castillo-Davis, C.; Meiklejohn, C.D.; Hartl, D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 2003, 300, 1742–1745. [Google Scholar] [CrossRef]
- Assis, R.; Zhou, Q.; Bachtrog, D. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 2012, 4, 1189–1200. [Google Scholar] [CrossRef]
- Perry, J.C.; Harrison, P.W.; Mank, J.E. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol. Biol. Evol. 2014, 31, 1206–1219. [Google Scholar] [CrossRef]
- Mank, J.E.; Nam, K.; Brunstrom, B.; Ellegren, H. Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol. Biol. Evol. 2010, 27, 1570–1578. [Google Scholar] [CrossRef]
- Pointer, M.A.; Harrison, P.W.; Wright, A.E.; Mank, J.E. Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genet. 2013, 9, e1003697. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; McNulty, S.; Meyskens, F.L., Jr. During human melanoma progression AP-1 binding pairs are altered with loss of c-Jun in vitro. Pigment Cell Res. 2004, 17, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Reinius, B.; Johansson, M.M.; Radomska, K.J.; Morrow, E.H.; Pandey, G.K.; Kanduri, C.; Sandberg, R.; Williams, R.W.; Jazin, E. Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genom. 2012, 13, 607. [Google Scholar] [CrossRef] [PubMed]
- Small, C.M.; Carney, G.E.; Mo, Q.; Vannucci, M.; Jones, A.G. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: Evidence for masculinization of the transcriptome. BMC Genom. 2009, 10, 579. [Google Scholar] [CrossRef] [PubMed]
- Parsch, J.; Ellegren, H. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013, 14, 83–87. [Google Scholar] [CrossRef]
- Meckbach, C.; Tacke, R.; Hua, X.; Waack, S.; Wingender, E.; Gültas, M. PC-TraFF: Identification of potentiallycollaborating transcription factors using pointwise mutual information. BMC Bioinform. 2015, 16, 400. [Google Scholar] [CrossRef] [PubMed]
- Meckbach, C.; Wingender, E.; Gültas, M. Removing Background Co-occurrences of Transcription Factor Binding Sites Greatly Improves the Prediction of Specific Transcription Factor Cooperations. Front. Genet. 2018, 9, 189. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]
- Klees, S.; Lange, T.M.; Bertram, H.; Rajavel, A.; Schlüter, J.S.; Lu, K.; Schmitt, A.O.; Gültas, M. In Silico Identification of the Complex Interplay between Regulatory SNPs, Transcription Factors, and Their Related Genes in Brassica napus L. Using Multi-Omics Data. Int. J. Mol. Sci. 2021, 22, 789. [Google Scholar] [CrossRef]
- Heinrich, F.; Wutke, M.; Das, P.P.; Kamp, M.; Gültas, M.; Link, W.; Schmitt, A.O. Identification of regulatory SNPs associated with vicine and convicine content of vicia faba based on genotyping by sequencing data using deep learning. Genes (Basel) 2020, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Triska, M.; Solovyev, V.; Baranova, A.; Kel, A.; Tatarinova, T.V. Nucleotide patterns aiding in prediction of eukaryotic promoters. PLoS ONE 2017, 12, e0187243. [Google Scholar] [CrossRef] [PubMed]
- Shahmuradov, I.A.; Umarov, R.K.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 2017, 45, e65. [Google Scholar] [CrossRef]
- Kumari, S.; Ware, D. Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots. PLoS ONE 2013, 8, e79011. [Google Scholar] [CrossRef]
- Molina, C.; Grotewold, E. Genome wide analysis of Arabidopsis core promoters. BMC Genom. 2005, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Wingender, E.; Chen, X.; Fricke, E.; Geffers, R.; Hehl, R.; Liebich, I. Match-a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2001, 29, 281–283. [Google Scholar] [CrossRef]
- Wingender, E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 2008, 9, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Heicklen-Klein, A.; McReynolds, L.J.; Evans, T. Using the zebrafish model to study GATA transcription factors. Semin. Cell Dev. Biol. 2005, 16, 95–106. [Google Scholar] [CrossRef]
- Holtzinger, A.; Evans, T. Gata4 regulates the formation of multiple organs. Development 2005, 132, 4005–4014. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.K.; Chung, B.C. Analysis of zebrafish cyp19 promoters. J. Steroid Biochem. Mol. Biol. 2003, 86, 381–386. [Google Scholar] [CrossRef]
- Takahashi, H. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 1977, 28, 57–65. [Google Scholar]
- Rodriguez-Mari, A.; Yan, Y.L.; Bremiller, R.A.; Wilson, C.; Canestro, C.; Postlethwait, J.H. Characterization and expression pattern of zebrafish Anti-Mullerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 2005, 5, 655–667. [Google Scholar] [CrossRef]
- Uchida, D.; Yamashita, M.; Kitano, T.; Iguchi, T. Oocyte apoptosis during the transition from ovarylike tissue to testes during sex differentiation of juvenile zebrafish. J. Exp. Biol. 2002, 205, 711–718. [Google Scholar] [CrossRef]
- Rodriguez-Mari, A.; Postlethwait, J.H. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 2011, 105, 461–490. [Google Scholar]
- Pradhan, A.; Khalaf, H.; Ochsner, S.A.; Sreenivasan, R.; Koskinen, J.; Karlsson, M.; Karlsson, J.; McKenna, N.J.; Orbán, L.; Olsson, P.E. Activation of NF-κB protein prevents the transition from juvenile ovary to testis and promotes ovarian development in zebrafish. J. Biol. Chem. 2012, 287, 37926–37938. [Google Scholar] [CrossRef]
- Sreenivasan, R.; Jiang, J.; Wang, X.; Bartfai, R.; Kwan, H.Y.; Christoffels, A.; Orban, L. Gonad differentiation in zebrafish is regulated by the canonical Wnt signaling pathway. Biol. Reprod. 2014, 90, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Slanchev, K.; Stebler, J.; de la Cueva-Mendez, G.; Raz, E. Development without germ cells: The role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 4074–4079. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Nüsslein-Volhard, C. Germ line control of female sex determination in zebrafish. Dev. Biol. 2008, 324, 277–287. [Google Scholar] [CrossRef]
- Pradhan, A.; Olsson, P.E. Regulation of zebrafish gonadal sex differentiation. AIMS Mol. Sci. 2016, 3, 567–584. [Google Scholar] [CrossRef]
- Von Hofsten, J.; Olsson, P.E. Zebrafish sex determination and differentiation: Involvement of FTZ-F1 genes. Reprod. Biol. Endocrinol. 2005, 3, 63. [Google Scholar] [CrossRef]
- Hebenstreit, D.; Wirnsberger, G.; Horejs-Hoeck, J.; Duschl, A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006, 17, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.; Gingras, S. Crucial role of cytokines in sex steroid formation in normal and tumoral tissues. Mol. Cell. Endocrinol. 2001, 171, 25–40. [Google Scholar] [CrossRef]
- Simard, J.; Ricketts, M.L.; Gingras, S.; Soucy, P.; Feltus, F.A.; Melner, M.H. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr. Rev. 2005, 26, 525–582. [Google Scholar] [CrossRef]
- Papacleovoulou, G.; Critchley, H.O.D.; Hillier, S.G.; Mason, J.I. IL1α and IL4 signalling in human ovarian surface epithelial cells. J. Endocrinol. 2011, 211, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Lachance, C.; Leclerc, P. Mediators of the Jak/STAT signaling pathway in human spermatozoa. Biol. Reprod. 2011, 85, 1222–1231. [Google Scholar] [CrossRef]
- Lin, J.C.; Hu, S.; Ho, P.H.; Hsu, H.J.; Postlethwait, J.H.; Chung, B.C. Two Zebrafish hsd3b Genes Are Distinct in Function, Expression, and Evolution. Endocrinology 2015, 156, 2854–2862. [Google Scholar] [CrossRef]
- Kuo, M.W.; Postlethwait, J.H.; Lee, W.C.; Lou, S.W.; Chan, W.K.; Chung, B.C. Gene duplication, gene loss and evolution of expression domains in the vertebrate nuclear receptor NR5A (Ftz-F1) family. Biochem. J. 2005, 389, 19–26. [Google Scholar] [CrossRef]
- Zheng, X.; Cheng, M.; Xiang, L.; Liang, J.; Xie, L.; Zhang, R. The AP-1 transcription factor homolog Pf-AP-1 activates transcription of multiple biomineral proteins and potentially participates in Pinctada fucata biomineralization. Sci. Rep. 2015, 5, 14408. [Google Scholar] [CrossRef]
- Martin, L.J.; Tremblay, J.J. The nuclear receptors NUR77 and SF1 play additive roles with c-JUN through distinct elements on the mouse Star promoter. J. Mol. Endocrinol. 2009, 42, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Francis Bergeron, F.; Viger, R.S.; Tremblay, J.J. Functional cooperation between GATA factors and cJUN on the star promoter in MA-10 Leydig cells. J. Androl. 2012, 33, 81–87. [Google Scholar] [CrossRef]
- Mooradian, A.D.; Morley, J.E.; Korenman, S.G. Biological actions of androgens. Endocr. Rev. 1987, 8, 1–28. [Google Scholar] [CrossRef]
- Kido, Y.; Sakazume, S.; Abe, Y.; Oto, Y.; Itabashi, H.; Shiraishi, M.; Yoshino, A.; Tanaka, Y.; Obata, K.; Murakami, N.; et al. Testosterone replacement therapy to improve secondary sexual characteristics and body composition without adverse behavioral problems in adult male patients with prader–willi syndrome: An observational study. Am. J. Med. Genet. A 2013, 161, 2167–2173. [Google Scholar] [CrossRef]
- Sekar, N.; Lavoie, H.A.; Veldhuis, J.D. Concerted regulation of steroidogenic acute regulatory gene expression by luteinizing hormone and insulin (or insulin-like growth factor I) in primary cultures of porcine granulosa-luteal cells. Endocrinology 2000, 141, 3983–3992. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Devoto, L.; Kohen, P.; Gonzalez, R.R.; Castro, O.; Retamales, I.; Vega, M.; Carvallo, P.; Christenson, L.K.; Strauss, J.F., III. Expression of steroidogenic acute regulatory protein in the human corpus luteum throughout the luteal phase. J. Clin. Endocrinol. Metab. 2001, 86, 5633–5639. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.R.; Eubank, D.W.; Stocco, D.M. Assessment of the role of activator protein-1 on transcription of the mouse steroidogenic acute regulatory protein gene. Mol. Endocrinol. 2004, 18, 558–573. [Google Scholar] [CrossRef]
- Clem, B.F.; Hudson, E.A.; Clark, B.J. Cyclic adenosine 30,50-monophosphate (cAMP) enhances cAMP-responsive element binding (CREB) protein phosphorylation and phospho-CREB interaction with the mouse steroidogenic acute regulatory protein gene promoter. Endocrinology 2005, 146, 1348–1356. [Google Scholar] [CrossRef][Green Version]
- Silverman, E.; Yivgi-Ohana, N.; Sher, N.; Bell, M.; Eimerl, S.; Orly, J. Transcriptional activation of the steroidogenic acute regulatory protein (StAR) gene: GATA-4 and CCAAT/enhancer-binding protein beta confer synergistic responsiveness in hormone-treated rat granulosa and HEK293 cell models. Mol. Cell. Endocrinol. 2006, 252, 92–101. [Google Scholar] [CrossRef]
- Lehoux, J.G.; Fleury, A.; Ducharme, L. The acute and chronic effects of adrenocorticotropin on the levels of messenger ribonucleic acid and protein of steroidogenic enzymes in rat adrenal in vivo. Endocrinology 1998, 139, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.R.; Stocco, D.M. The role of JUN in the regulation of PRKCC-mediated STAR expression and steroidogenesis in mouse Leydig cells. J. Mol. Endocrinol. 2008, 41, 329–341. [Google Scholar] [CrossRef]
- Silverman, E.; Eimerl, S.; Orly, J. CCAAT enhancer-binding protein beta and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J. Biol. Chem. 1999, 274, 17987–17996. [Google Scholar] [CrossRef] [PubMed]
- LaVoie, H.A.; Singh, D.; Hui, Y.Y. Concerted regulation of the porcine StAR gene promoter activity by FSH and IGF-I in granulosa cells involves GATA-4 and C/EBPb. Endocrinology 2004, 145, 3122–3134. [Google Scholar] [CrossRef]
- Martin, L.J.; Taniguchi, H.; Robert, N.M.; Simard, J.; Tremblay, J.J.; Viger, R.S. GATA factors and the nuclear receptors, steroidogenic factor 1/liver receptor homolog 1, are key mutual partners in the regulation of the human 3beta-hydroxysteroid dehydrogenase type 2 promoter. Mol. Endocrinol. 2005, 19, 2358–2370. [Google Scholar] [CrossRef]
- Dube, C.; Bergeron, F.; Vaillant, M.J.; Robert, N.M.; Brousseau, C.; Tremblay, J.J. The nuclear receptors SF-1 and LRH-1 are expressed in endometrial cancer cells and regulate steroidogenic gene transcription by cooperating with AP-1 factors. Cancer Lett. 2009, 275, 127–138. [Google Scholar] [CrossRef]
- Bauer, M.P.; Bridgham, J.T.; Langenau, D.M.; Johnson, A.L.; Goetz, F.W. Conservation of steroidogenic acute regulatory (StAR) protein structure and expression in vertebrates. Mol. Cell. Endocrinol. 2000, 168, 119–125. [Google Scholar] [CrossRef]
- Ings, J.S.; Kraak, G.J.V.D. Characterization of the mRNA expression of StAR and steroidogenic enzymes in zebrafish ovarian follicles. Mol. Reprod. Dev. 2006, 73, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Ghouili, F.; Roumaud, P.; Martin, L.J. Gja1 expression is regulated by cooperation between SOX8/SOX9 and cJUN transcription factors in TM4 and 15P-1 Sertoli cell lines. Mol. Reprod. Dev. 2018, 85, 875–886. [Google Scholar] [CrossRef]
- Zhang, S.S.M.; Wei, J.Y.; Li, C.; Barnstable, C.J.; Fu, X.Y. Expression and activation of STAT proteins during mouse retina development. Exp. Eye Res. 2003, 76, 421–431. [Google Scholar] [CrossRef]
- Klein, R.; Klein, B.P.; Linton, K.L.P. Prevalence of agerelated maculopathy: The beaver eye study. Ophthalmology 1999, 299, 933–943. [Google Scholar]
- Del Priore, L.V.; Tezel, T.H.; Kaplan, H.J. Survival of allogeneic porcine retinal pigment epithelial sheets after subretinal transplantation. Investig. Ophthalmol. Vis. Sci. 2004, 45, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Valtinik, M.; Engelmann, K. Culturing of retinal pigment epithelium cells. Dev. Ophthalmol. 2009, 43, 109–119. [Google Scholar]
- Fasler-Kan, E.; Barteneva, N.S.; Ketterer, S.; Wunderlich, K.; Reschner, A.; Nurzhanova, A.; Flammer, J.; Huwyler, J.; Meyer, P. Human cytokines activate JAK-STAT signaling pathway in porcine ocular tissue. Xenotransplantation 2013, 20, 469–480. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rezai, K.A.; Kohen, L.; Wiedermann, P.; Heimann, K. Iris pigment epithelium transplantation. Graefes Arch. Clin. Exp. Ophthalmol. 1997, 235, 558–562. [Google Scholar] [CrossRef]
- Melville, H.; Carpiniello, M.; Hollis, K.; Staffaroni, A.; Golestaneli, N. Stem cells: A new paradigm for disease modeling and developing therapies for age-related macular degeneration. J. Transl. Med. 2013, 11, 53. [Google Scholar] [CrossRef]
- Choi, H.; Choi, H.; Han, J.; Jin, S.H.; Park, J.Y.; Shin, D.W.; Lee, T.R.; Kim, K.; Lee, A.-Y.; Noh, M. IL-4 inhibits the melanogenesis of normal human melanocytes through the JAK2-STAT6 signaling pathway. J. Investig. Dermatol. 2013, 133, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Chen, J.; Lu, J.; Yi, L.; Tong, X.; Kang, L.; Pei, S.; Ouyang, Y.; Jiang, L.; Ding, Y.; et al. Roles of inflammation factors in melanogenesis (Review). Mol. Med. Rep. 2020, 21, 1421–1430. [Google Scholar] [CrossRef]
- Fuchs, J.; Kern, H. Modulation of UV-light-induced skin inflammation by D-alpha-tocopherol and L-ascorbic acid: A clinical study using solar simulated radiation. Free Radic. Biol. Med. 1998, 25, 1006–1012. [Google Scholar] [CrossRef]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef]
- Martin, S.F. contact dermatitis: From pathomechanisms to immunotoxicology. Exp. Dermatol. 2012, 21, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Basler, K.; Brandner, J.M. Tight junctions in skin inflammation. Pflug. Arch. 2017, 469, 3–14. [Google Scholar] [CrossRef]
- Praetorius, C.; Grill, C.; Stacey, S.N.; Metcalf, A.M.; Gorkin, D.U.; Robinson, K.C.; Van Otterloo, E.; Kim, R.S.; Bergsteinsdottir, K.; Ogmundsdottir, M.H.; et al. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell 2013, 155, 1022–1033. [Google Scholar] [CrossRef]
- Visser, M.; Palstra, R.J.; Kayser, M. Allele-specific transcriptional regulation of IRF4 in melanocytes is mediated by chromatin looping of the intronic rs12203592 enhancer to the IRF4 promoter. Hum. Mol. Genet. 2015, 24, 2649–2661. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, Y.; Yong, H.X.L.; Fane, M.E.; Soogrim, A.; Lim, W.; Mahiuddin, D.N.; Kim, R.S.; Ashcroft, M.; Beatson, S.A.; Ainger, S.A.; et al. Genetic variation in IRF4 expression modulates growth characteristics, tyrosinase expression and interferon-gamma response in melanocytic cells. Pigment Cell Melanoma Res. 2018, 31, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Van Otterloo, E.; Li, W.; Bonde, G.; Day, K.M.; Hsu, M.Y.; Cornell, R.A. Differentiation of zebrafish melanophores depends on transcription factors AP2 alpha and AP2 epsilon. PLoS Genet. 2010, 6, e1001122. [Google Scholar] [CrossRef] [PubMed]
- Angel, P.; Szabowski, A.; Schorpp-Kistner, M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 2001, 20, 2413–2423. [Google Scholar] [CrossRef]
- Frantz, W.T.; Ceol, C.J. From tank to treatment: Modeling melanoma in zebrafish. Cells 2020, 9, 1289. [Google Scholar] [CrossRef]
- Heater, S.J.; Rains, J.D.; Braden, A.R.C.; Gilmer, S.M.; Walter, R.B. Cloning of JunA and JunB and comparison of mRNA expression levels in two Xiphophorus melanoma models. Zebrafish 2006, 3, 53–63. [Google Scholar] [CrossRef]
- Fan, R.; Xie, J.; Bai, J.; Wang, H.; Tian, X.; Bai, R.; Jia, X.; Yang, L.; Song, Y.; Herrid, M.; et al. Skin transcriptome profiles associated with coat color in sheep. BMC Genom. 2013, 14, 389. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.C.; Lamoreux, M.L. The color loci of mice—A genetic century. Pigment Cell Res. 2003, 16, 333–444. [Google Scholar] [CrossRef] [PubMed]
- Commo, S.; Gaillard, O.; Bernard, B. Human hair greying is linked to a specific depletion of hair follicle melanocytes affecting both the bulb and the outer root sheath. Br. J. Dermatol. 2004, 150, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Dong, Y.; Cao, J.; Bai, R.; Zhu, Z.; Li, P.; Zhang, J.; He, X.; Lü, L.; Yao, J.; et al. Gene expression profile in white alpaca (Vicugna pacos) skin. Animal 2011, 5, 1157–1161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cecchi, T.; Valbonesi, A.; Passamonti, P.; Frank, E.; Renieri, C. Quantitative variation of melanins in llama (Lama glama L.). Small Rumin. Res. 2007, 71, 52–58. [Google Scholar] [CrossRef]
- Sponenberg, D.P.; Ito, S.; Eng, L.A.; Schwink, K. Pigment types of various color genotypes of horses. Pigment Cell Res. 1988, 1, 410–413. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini, S.; Schmitt, A.O.; Tetens, J.; Brenig, B.; Simianer, H.; Sharifi, A.R.; Gültas, M. In Silico Prediction of Transcription Factor Collaborations Underlying Phenotypic Sexual Dimorphism in Zebrafish (Danio rerio). Genes 2021, 12, 873. https://doi.org/10.3390/genes12060873
Hosseini S, Schmitt AO, Tetens J, Brenig B, Simianer H, Sharifi AR, Gültas M. In Silico Prediction of Transcription Factor Collaborations Underlying Phenotypic Sexual Dimorphism in Zebrafish (Danio rerio). Genes. 2021; 12(6):873. https://doi.org/10.3390/genes12060873
Chicago/Turabian StyleHosseini, Shahrbanou, Armin Otto Schmitt, Jens Tetens, Bertram Brenig, Henner Simianer, Ahmad Reza Sharifi, and Mehmet Gültas. 2021. "In Silico Prediction of Transcription Factor Collaborations Underlying Phenotypic Sexual Dimorphism in Zebrafish (Danio rerio)" Genes 12, no. 6: 873. https://doi.org/10.3390/genes12060873
APA StyleHosseini, S., Schmitt, A. O., Tetens, J., Brenig, B., Simianer, H., Sharifi, A. R., & Gültas, M. (2021). In Silico Prediction of Transcription Factor Collaborations Underlying Phenotypic Sexual Dimorphism in Zebrafish (Danio rerio). Genes, 12(6), 873. https://doi.org/10.3390/genes12060873





